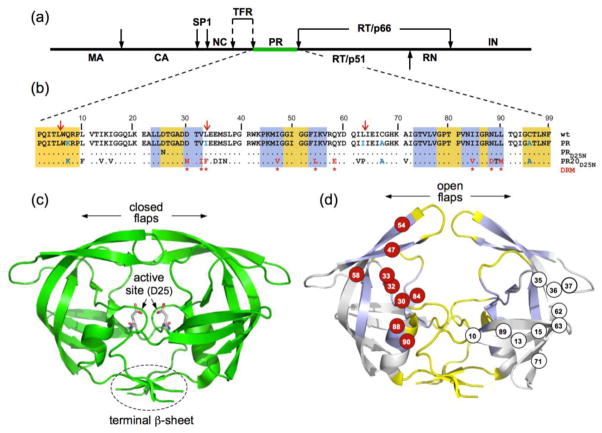
Figure 1.
(a) Domain organization of the HIV-1 Gag-Pol polyprotein. Abbreviations: MA, matrix; CA, capsid; SP1, spacer peptide 1; NC, nucleocapsid; TFR, transframe region; PR, protease; RT, reverse transcriptase; RN, ribonuclease; IN, integrase; (b) Sequence alignment of mature proteases used in this study. A construct optimized for structural studies and a surrogate of the wild-type, namely PR, bears mutations Q7K, L33I, L63I, C67A, and C95A (shown in blue) to restrict autoproteolysis (red arrows) and avoid cysteine-thiol oxidation45. Introduced mutations common to PR and PR20 are Q7K, C67A, and C95A. The 19 mutations, both naturally occurring and selected under drug pressure, in PR20 are shown in black and red lettering, respectively. Dots denote identical residues. Highly conserved regions and regions where major drug-resistance mutations occur are highlighted in yellow and light blue, respectively46. Regions of natural variation are not highlighted. Major drug-resistance mutations are indicted by red asterisks (as defined in http:/hivdb.stanford.edu/cgi-bin/PIResiNote.cgi and reference 47). X-ray structure of (c) the optimized wild-type protease (PR) in a closed conformation (PDB id: 3BVB)26 with active site residues shown in stick representation and (d) the inhibitor-free PR20 dimer in a wide-open conformation (PDB ID: 3UF3)15. (d) The location of major drug-resistance mutations (DRMs) are shown on one subunit in red circles and naturally selected/compensatory mutations are shown on the other subunit in black on white circles. Highly conserved regions and regions where major drug-resistance mutations occur are shaded on the x-ray structure in yellow and light blue, respectively, matching the color scheme in (b) for those same regions.