Figure 1.
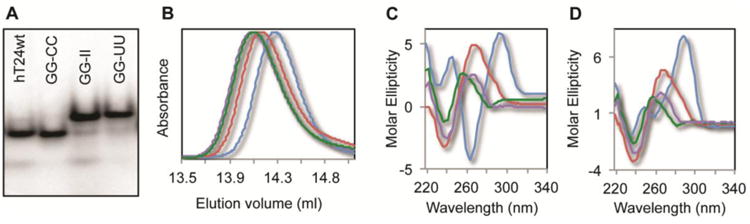
Secondary structure analysis of DNA substrates. (A) Nondenaturing gel electrophoresis reveals different mobility of the various DNA sequences. hT24wt and hT24GG-CC migrate faster, which is indicative of more compact folded structures of these DNA substrates. Conversely, hT24GG-II and hT24GG-UU, which have no internal secondary structure, migrate slower in the gel. (B) Normalized chromatograms of the 24-nt DNA substrates investigated using SE-HPLC in NaCl buffer. hT24wt (blue line) elutes slowest, followed by hT24GG-CC (red line), and hT24GG-II (green line) and hT24GG-UU (purple line) eluting first. Size-exclusion chromatography separates DNA substrates based on shape, with more compact structures eluting later than those that are less compact. CD spectra of hT24wt, hT24GG-CC, hT24GG-II, and hT24GG-UU in (C) Na+ or (D) K+ containing buffer. hT24wt shows a spectrum consistent with antiparallel topology in Na+ ions and hybrid parallel/antiparallel topology in K+ ions. Spectra for hT24GG-CC, hT24GG-II, and hT24GG-UU are similar in Na+ and K+ ions, indicating that their structures are not significantly altered in different monovalent ion solutions. Colors of lines are the same as those in panel (B).
