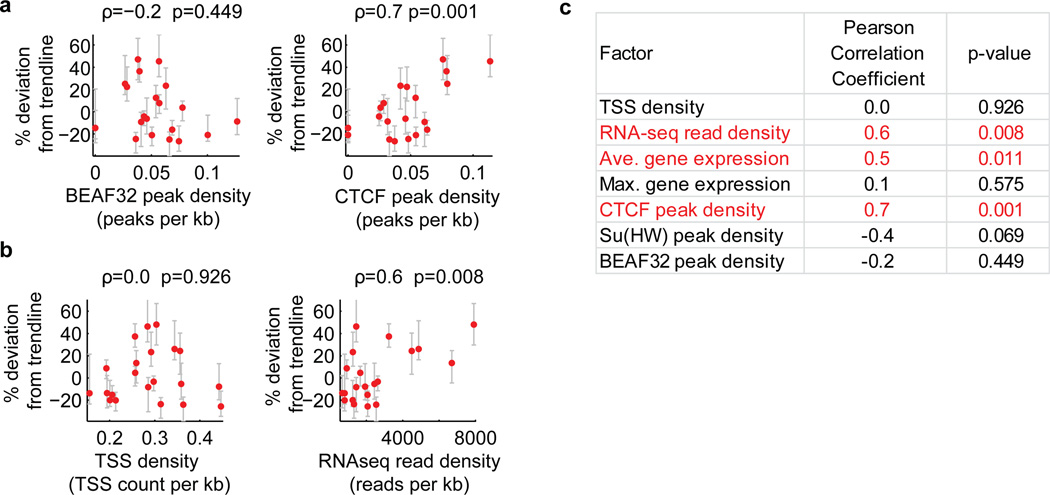
Extended Data Figure 9. Additional factors correlating with the domain volume after normalizing the effect of domain length for active domains.
To normalize for the effect of domain length, we determined the percent deviation of the volumes of active domains from the power law scaling trendline shown in Fig. 1c, and hereafter refer to this value as percent deviation from trendline. a, Correlation of the percent deviation from trendline with the binding density of the insulator proteins BEAF32 (left) and CTCF (right). Binding density was determined from the density of peaks per kb in Dam-ID data20. Peaks were defined as local maxima at least 2 standards deviation above the mean. b, As in (a) but for correlation with transcription start site (TSS) density (left) and RNA-seq total read density (right). The TSS density is defined as the average number of TSSs per kb in the domain and the RNA-seq total read density is defined as the total number of reads mapping to the domain measured using RNA sequencing divided by the domain length in kb. c, Pearson Correlation coefficients and corresponding p-values for the correlation of percent deviation from trendline with the indicated genomic factors. Average gene expression is referring to the average expression value in FPKM (read fragments per kilobase per million reads) of all genes in the domain. Maximum gene expression is referring to the FPKM of the most highly expressed gene in the domain. Su(Hw) is an insulator protein like BEAF32 and CTCF. We noticed a weak trend in which domains with higher binding densities of the insulators BEAF32 or Su(Hw) are slightly more compact. Although this trend is consistent with the hypothesis that insulator proteins may function as loop forming factors and that loops may lead to more compact domains12, the correlation detected here was not statistically significant. Further analysis with improved sensitivity in detection of BEAF32 or Su(Hw) binding sites might uncover a stronger affect, so our data do not rule out the insulator loop hypothesis. Similarly, we caution that the positive correlation observed with the density of CTCF binding sites might reflect the preference of CTCF to bind open chromatin regions (such as enhancers and promoters), and does not necessarily suggest that CTCF binding induces a more open chromatin state.