FIGURE 1.
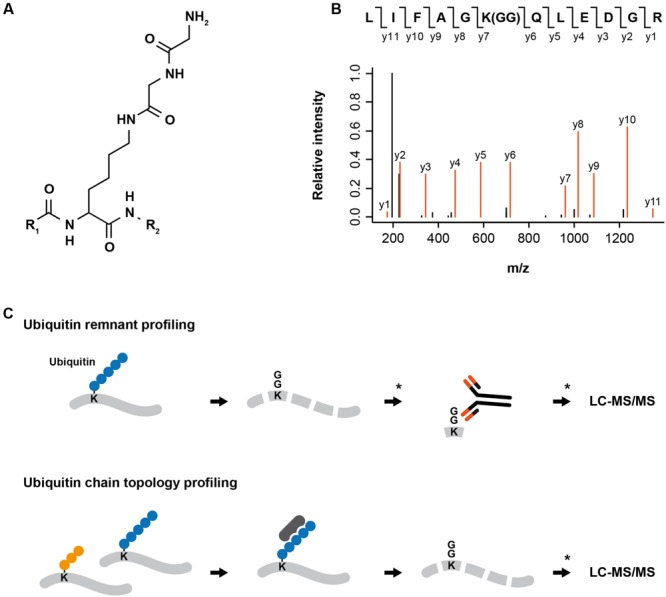
MS-based proteomics approaches for analyzing protein ubiquitylation. (A) Digestion of ubiquitylated proteins with trypsin leaves a di-glycine remnant from the C-terminus of ubiquitin covalently attached to the previously modified lysine. (B) Exemplary fragment spectrum of a di-glycine modified peptide. The di-glycine remnant leads to a shift of ∼114 Da in the peptide mass and can be exploited to pinpoint the localization of the ubiquitin attachment. (C) For ubiquitin remnant profiling, proteins extracted from cells are digested into peptides using trypsin, di-glycine modified peptides are enriched using di-glycine lysine specific antibodies and identified by LC-MS/MS. ∗Pre- and post-enrichment fractionation can be introduced to decrease sample complexity and increase the depth of the analysis. For ubiquitin chain topology profiling, proteins extracted from cells or tissues are incubated with an ubiquitin linkage-specific binder (e.g., antibody, affimer, TUBE). Enriched proteins modified by a specific type of ubiquitin chain are digested in-gel into peptides and peptide samples are analyzed by LC-MS/MS. ∗Post-enrichment fractionation can be introduced to decrease sample complexity and increase the depth of the analysis.
