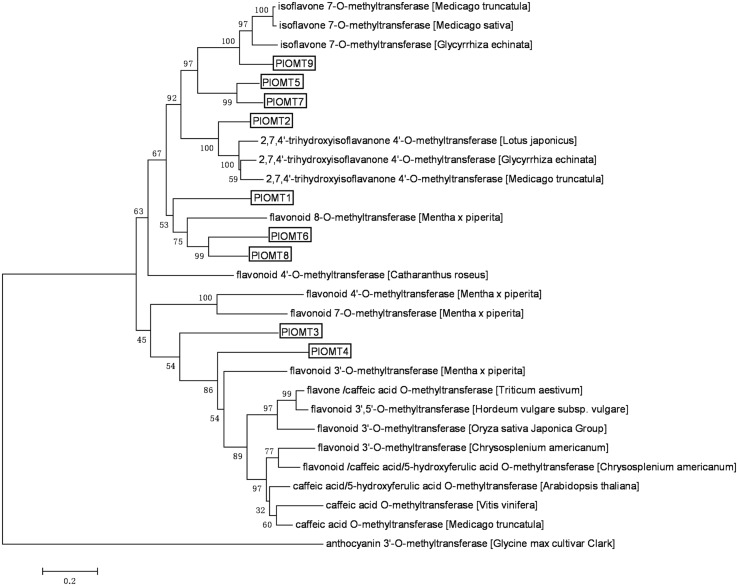
FIGURE 2.
Phylogenetic analysis of the PlOMTs with other OMTs whose functions are already known. Amino acids were aligned using the CLUSTAL X Version 2.0 program, and the phylogenetic tree was constructed by the neighbor-joining method of MEGA 6.0. Numbers on each node indicate the bootstrap values of 1000 replicates. The scale bar represents 0.2 amino acid substitutions per site.