Abstract
Betaproteobacteria were the most common isolates from the water‐filled tank of a Costa Rican bromeliad. Isolates included eight species from the orders Neisseriales and Burkholderiales, with close relatives recovered previously from tropical soils, wetlands, freshwater, or in association with plants. Compared to close relatives, the isolates displayed high temperature and comparatively low pH optima, reflecting the tropical, acidic nature of the bromeliad tank. Bromeliad‐associated bacteria most closely related to Chromobacterium, Herbaspirillum, and Aquitalea were all isolated exclusively at pH 6, while Ralstonia, Cupriavidus, and three species of Burkholderia were isolated mostly at pH 4. Activity profiles for the isolates suggest pervasive capabilities for the breakdown of plant‐sourced organics, including d‐galacturonic acid, mannitol, d‐xylose, and l‐phenylalanine, also reflecting a niche dominated by decomposition of leaves from the overlying canopy, which become entrained in the tanks. Metabolic activity profiles were overlapping between the Burkholderiales, isolated at pH 4, and the Neisseriales, isolated at pH 6, suggesting that plant material decomposition, which is presumably the underlying process sustaining the tank community and possibly the plant itself, occurs in the tanks at both pH extremes. These results suggest that bromeliad‐associated betaproteobacteria may play an important role in the cycling of carbon in this unusual aquatic habitat.
Keywords: Betaproteobacteria, bromeliad, Costa Rica, decomposition, rainforest
Introduction
Plants from the family Bromeliaceae are prominent members of neotropical rain and cloud forests. Bromeliad average density has been estimated at 1000–100,000 ha−1 ground area, depending on the study (Sugden and Robins 1979; Richardson 1999). Many species collect large amounts of water in the canopy by the formation of unique foliar arrangements, or “tanks” (Richardson 1999; Benzing 2000), thus these densities represent as much as 50,000 L suspended water in the canopy ha−1 (Fish 1983). This creates an unusual aquatic habitat suspended in the canopy, allowing for long‐term retention and decomposition of organic compounds, compared to soil (Pittl et al. 2010; Goffredi et al. 2011a,b). The physiological capabilities of bromeliads, including increased capacity for amino acid and mineral uptake from the tank, are known (Benzing 1970; Winkler and Zotz 2009), but the specific metabolisms of resident bacteria, and their role in carbon cycling in the rainforest, have not been specifically elucidated.
Several studies have investigated the diversity and certain processes driven by microbes within bromeliad tanks. Bacterial abundance has been shown to be 2–5× higher in bromeliad tank debris, compared to ground soils (Pittl et al. 2010) and tank habitat heterogeneity supports a myriad of bacterial residents, including at least 21 bacterial orders/subdivisions, many of which are closely related to bacteria previously found in soil, peat bogs, and stagnant water (Dedysh et al. 2006; Inselsbacher et al. 2007; Rui et al. 2009; Yarwood et al. 2009; Goffredi et al. 2011a). Bromeliad tanks are highly stratified and within a single tank can range from 4 to 6 pH and from 0.5 to 8.0 ppm O2, providing many possible niches available within these unusual microbial ecosystems (Laessle 1961; Benzing et al. 1972; Guimaraes‐Souza et al. 2006; Goffredi et al. 2011a,b). Tank pH, in particular, is a major influence on the resident microbial population. In tanks of pH <5, the bacterial community was dominated by Acidobacteria and Alpha‐proteobacteria, while tanks of pH >5 were dominated by Firmicutes and Betaproteobacteria (Goffredi et al. 2011a).
Given the unique nature of the bromeliad tank habitat and the ecological importance of bromeliads in general, it is compelling to further examine specific associated bacterial community members and the possible influences they may have on nutrient cycling. To that end, this study sought to examine the specific capabilities of bromeliad‐associated bacteria, with emphasis on heterotrophic degradation of organic matter, and environmental tolerances. Eight bacterial representatives from three major families within the betaproteobacteria were isolated and characterized, taxonomically and physiologically, from a tank water sample collected from a Costa Rican bromeliad (Werauhia gladioliflora). It is expected that these, and other, bromeliad‐associated bacteria play important roles in cycling recalcitrant carbon in the rainforest.
Methods
Isolation
Tank water, with an initial pH of 5.0, was obtained from a bromeliad (Werauhia gladioliflora) at La Selva Biological Research Station, Costa Rica, in January 2012. Bacteria were cultivated by streaking serial dilutions on Standard Methods Agar (SMA) of pH 4 or 6. Plates were incubated at 30°C for 1–3 days under aerobic conditions. Single colonies were selected based on unique morphological characteristics (Fig. 1) and further purified by standard T‐streak. The number of viable bacteria in the starting sample was estimated by plating serial dilutions (10−1, 10−2, and 10−3) on Nutrient Agar. Colony‐forming units were counted following incubation for 3 day at 30°C.
Figure 1.
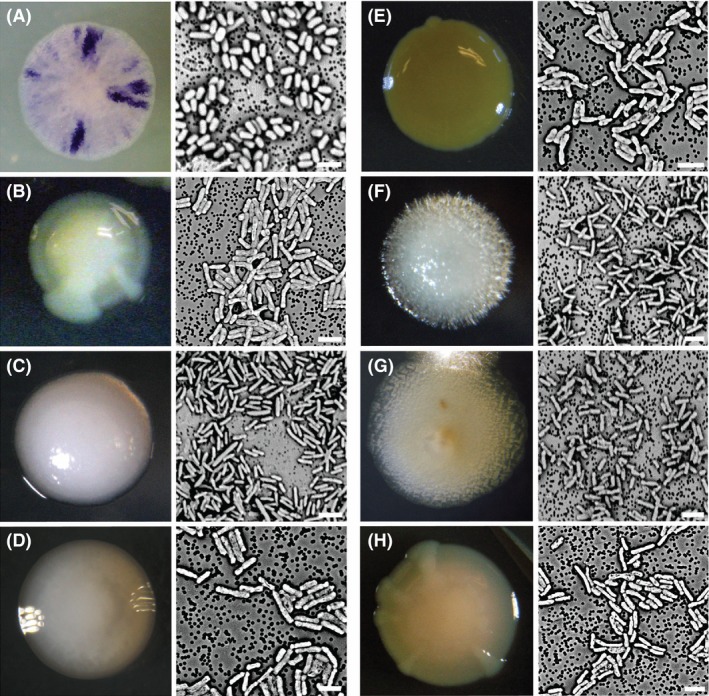
Colony and cell morphology of bromeliad‐associated bacterial isolates. (A) Isolate Br4, related to Chromobacterium. (B) Isolate Br23, related to Aquitalea. (C) Isolate Br2, related to Cupriavidus. (D) Isolate Br27, related to Ralstonia. (E) Isolate Br3, related to Burkholderia. (F) Isolate Br19, related to Burkholderia. (G) Isolate Br6, related to Burkholderia. (H) Isolate Br24, related to Herbaspirillum. Image at left of each letter is the colony morphology at 7 days of growth. Image at right of each letter is the individual cell morphology taken via scanning electron microscopy. The specific scales were not originally noted for colony sizes. Bars for the SEM images, 2 μm.
Morphology
Individual cell morphology was determined via scanning electron microscopy (SEM; Fig. 1). Bacterial samples for SEM were initially fixed in 3% glutaraldehyde in 0.1mol L‐1 cacodylate for 72 h at 4°C. Samples were then pulled onto a 0.22 μm polycarbonate filter (Millipore, Billerica, MA), washed in a graded ethanol series (50%, 75%, and 100%) and placed in hexamethyldisilazane for 1 h at room temperature. Filters were then mounted, palladium‐coated (Hummer VI, Union City, CA), and visualized using a Phenom desktop SEM (FEI Instruments, Hillsboro, OR).
DNA extraction, PCR amplification, and 16S rRNA sequencing
Genomic DNA from each isolate was extracted using the Qiagen DNeasy kit (Qiagen, CA) according to the manufacturer's instruction. 16S rRNA genes were amplified using primers 27F and 1492R, according to Lane 1991; Successful 16S rRNA gene products were cleaned with MultiScreen HTS plates (Millipore Corporation) and sequenced at Laragen, Inc. (Culver City, CA). Closest relatives were identified using BLASTn (Altschul et al. 1997). Sequences were assembled, edited, and aligned using Sequencher v4.10.1 (GeneCodes Corp, Ann Arbor, MI). For bromeliad isolates and closest relatives, neighbor‐joining analysis was used to show phylogenetic relationships (Fig. 2), while maximum parsimony analysis was performed using the heuristic search option with 1000 bootstrap replicates to assign confidence levels to nodes (using PAUP*4.0b10; Swofford 1998). GenBank accession numbers for 16S rRNA sequences obtained in this study are KT387842–KT387849.
Figure 2.
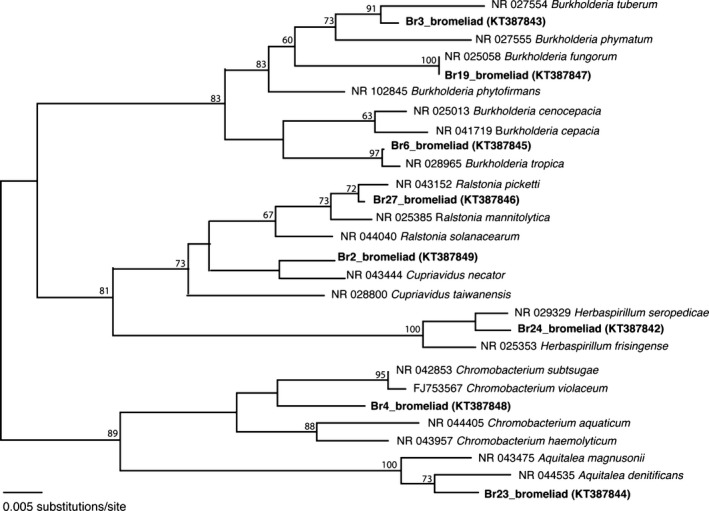
Phylogenetic relationships among betaproteobacteria associated with a Costa Rican bromeliad, relative to selected cultured representatives in public databases, based on sequence divergence within the 16S rRNA gene. (A) neighbor‐joining tree with Kimura two‐parameter distances is shown. The numbers at the nodes represent maximum parsimony bootstrap values from 1000 replicate samplings (only values >60% are shown).
Metabolic characteristics
Amylase function was determined by streak of bacterial isolates on agar plates enriched with starch. After 2 days at 30°C, iodine was added and a zone of clearing was observed. Protein hydrolysis was assessed, via a zone of clearing, on a skim milk plate containing casein. Additional carbon assimilation capabilities were tested using EcoPlates (Biolog Inc., Hayward, CA). Arrays were set up according to manufacturer's instruction using a bacterial suspension with 90% transmissivity, as determined by a Spectronic 20 spectrometer (Milton Roy Company, Warminster, PA). Biolog EcoPlates were incubated at 30°C in the dark and measured intermittently for 12–60 h for changes in absorbance at 590 nm, using an EL800 Microplate Reader (Bio‐Tek Instruments, Winooski, VT). Two statistical comparisons of phenotype patterns among isolates and strains were performed. A hierarchical cluster analysis of bacterial phenotypes, based on Bray–Curtis similarity resemblance (presence/absence, single linkage) of the sum of 19 carbon substrate utilization capabilities, was conducted using Primer v6 (Fig. 4; Clarke and Gorley 2006). Additionally, a Z‐test for two population proportions was conducted to determine significance between specific growth and metabolic capabilities of the bromeliad‐associated isolates (assigned as population 1) and closely related strains (assigned as population 2; Table 1).
Figure 4.
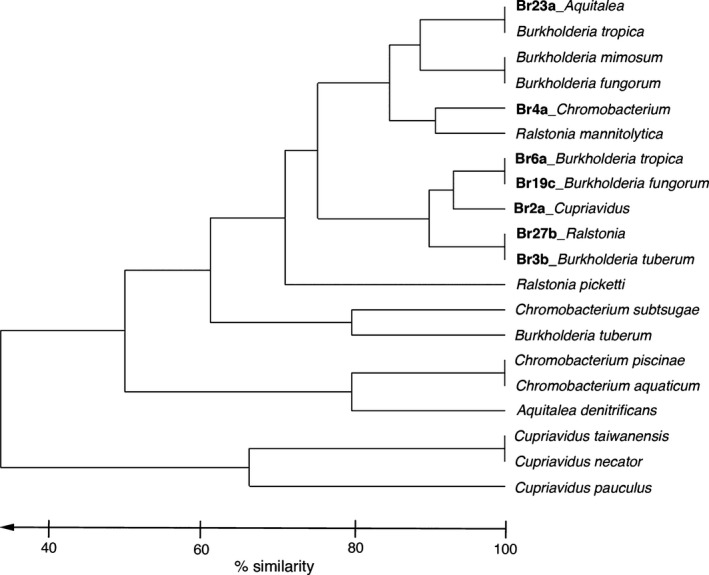
Hierarchical cluster analysis of metabolic phenotypes for bromeliad‐associated isolates recovered in this study, versus strains detailed in other publications. Phenotypic character traits included 19 carbon substrate utilization capabilities, revealed by Biolog EcoPlates, shown in Fig. 3, The analysis, based on Bray–Curtis similarity resemblance (presence/absence, single linkage), was performed using Primer v6.
Table 1.
Select growth conditions and phenotypic characteristics of bromeliad tank isolates in comparison to other closely related strains
| Isolate/Strain | 37°C | 42°C | pH 4 | pH 5 | Mannitol | l‐phenyl | Xylose |
|---|---|---|---|---|---|---|---|
| Br3 | + | + | + | + | + | + | — |
| Br19 | + | + | + | + | + | + | + |
| Br6 | + | + | + | + | + | + | + |
| Br2 | + | + | + | + | + | + | + |
| Br27 | + | + | + | + | + | + | — |
| Br4 | + | + | − | + | + | + | + |
| Br23 | + | − | − | + | + | + | + |
| B. tuberum | + | − | na | na | + | na | na |
| B. fungorum | + | − | na | na | + | na | + |
| B. tropica | + | − | − | + | + | + | + |
| B. mimosarum | + | − | na | na | + | + | + |
| C. necator | + | − | − | − | − | na | − |
| C. pauculus | + | + | na | na | − | na | − |
| C. taiwanensis | + | − | na | na | − | na | − |
| R. picketti | + | + | na | na | − | + | + |
| R. mannitolytica | + | + | na | na | + | + | + |
| C. subtsugae | + | − | − | − | − | − | − |
| C. aquaticum | − | − | − | + | − | + | − |
| C. violaceum | − | − | − | − | − | − | na |
| C. piscinae | − | − | − | + | − | − | − |
| A. denitrificans | + | − | − | + | − | − | na |
| Isolate proportion | 1.00 | 0.86 | 0.71 | 1.00 | 1.00 | 1.00 | 0.71 |
| Strain proportiona | 0.79 | 0.21 | 0.00 | 0.57 | 0.36 | 0.56 | 0.45 |
| Z‐score | 1.323 | 2.806 | 2.789 | 1.954 | 2.806 | 2.037 | 1.081 |
| P valueb | 0.1868 | 0.0049 | 0.0053 | 0.0512 | 0.0049 | 0.0414 | 0.2801 |
Strain references: Burkholderia tuberum (Vandamme et al. 2002). B. tropica (Reis et al. 2004; Aizawa et al. 2010), B. mimosarum (Chen et al. 2006), B. fungorum (Coenye et al. 2001), B. phytofirmans (Sessitsch et al. 2005). Cupriavidus necator (Makkar and Casida 1987); C. pauculus (Vandamme et al. 1999; Vaneechoutte et al. 2004); C. taiwanensis (Chen et al. 2001; Vaneechoutte et al. 2004); Ralstonia picketti (Coenye et al. 2003); R. mannitolytica (De Baere et al. 2001; Coenye et al. 2003). Chromobacterium subtsugae (Martin et al. 2007); C. aquaticum (Young et al. 2008; Kampfer et al. 2009); C. violaceum (Martin et al. 2007; Young et al. 2008); C. piscinae (Kampfer et al. 2009), Aquitalea denitrificans (Lee et al. 2009).
Proportion calculated for only those strains with known values.
Based on Z‐test for 2 population proportions, with two‐tailed hypothesis.
l‐phenyl, l‐phenylalanine; na, not available from previous studies. bold values denote significance with P values < 0.0512
Temperature and pH tolerance studies
The temperature ranges for growth were determined visually by 5‐day incubations at 23, 30, 37, 42, and 47°C on SMA plates. The pH ranges were determined via 5‐day incubations at 30°C in Tryptic Soy Broth of incremental pH values between 4 and 7.5, adjusted with 1N HCl or 1M NaOH. Growth was determined via decreases in transmissivity of the suspension, as measured via a Spectronic 20 spectrometer at OD600 (Milton Roy Company, Houston, TX).
Results
Identity of bromeliad‐associated bacterial isolates
The original Costa Rican bromeliad (Werauhia gladioliflora) tank water sample had a total aerobic microbial cell count of 3.2 × 105 c.f.u. mL−1 of tank water. Thirty‐five of 41 isolates recovered were members of the betaproteobacteria, five were members of the genus Bacillus (Firmicutes) and one was a member of the genus Roseomonas (Alpha‐proteobacteria). Betaprotoeobacteria isolates comprised eight distinct types (with representatives shown in Fig. 1). Their phylogenetic relationships are shown in Figure 2; three members of the genus Burkholderia (order Burkholderiales) most closely related to B. tuberum (Br3, 98% similarity in 16S rRNA), B. fungorum (Br19, 100% similarity), and B. tropica (Br6, 99% similarity), one closely related to Cupriavidus necator (Br2, 97% similarity in 16S rRNA; order Burkholderiales), one most closely related to a species within the sister genus, Ralstonia picketti (Br27, 99% similarity, order Burkholderiales), one most closely related to Herbaspirillum seropedicae (Br24, 99% similarity in 16S rRNA, order Burkholderiales), one most closely related to Chromobacterium subtsugae (Br4, 99% similarity in 16S rRNA; order Neisseriales), and one isolate was most closely related to Aquitalea denitrificans (Br23, 99% similarity, order Neisseriales).
Phenotypic characterization of bromeliad‐associated bacterial isolates
All isolates were observed to grow at temperatures ranging from 23 to 42°C (Tables S1–S3), with the exception of one isolate Br2, related to Cupriavidus, which grew at 47°C. Growth at 42°C was a significantly distinct feature of the bromeliad‐associated isolates (P = 0.0049; Table 1), compared to related strains. In general, the Burkholderiales were isolated at pH 4 (and grew well at a range of 4–7), while the Neisseriales were isolated at pH 6, and grew only above pH 5 (Tables S1–3). Similarly, the ability to grow at pH 4 and 5 was more common for the bromeliad‐associated isolates (P = 0.0053 and 0.0512, respectively; Table 1), than for related strains.
Isolates of each betaproteobacteria type (n = 1–8) were surveyed for their metabolic capabilities (Fig. 3; Tables S1–3). Metabolic activity profiles were overlapping between the Burkholderiales, isolated at pH 4, and the Neisseriales, isolated at pH 6 (Fig. 3). EcoPlate assays revealed the positive use of the following carbon sources by all eight isolate types: d‐galacturonic acid, l‐asparagine, d‐mannitol, N‐acetyl‐d‐glucosamine, and d‐glucosaminic Acid (Fig. 3; Tables S1–3). At least six isolate types demonstrated use of pyruvic acid methyl ester, d‐xylose, l‐threonine, and l‐phenylalanine. Fewer than five isolate types utilized Tween 40, Tween 80, l‐serine, and d‐malic acid (Fig. 3; Tables S1–3). The use of mannitol and l‐phenylalanine were both significantly distinct features of the bromeliad‐associated isolates (P = 0.0049 and 0.0414, respectively; Table 1), compared to related strains. Hierarchical cluster analysis revealed that bromeliad‐associated isolates were generally distinct from their closest relatives, with regard to carbon utilization profiles (the collective ability to use any of 19 carbon substrates shown in Fig. 3 and 4).
Figure 3.
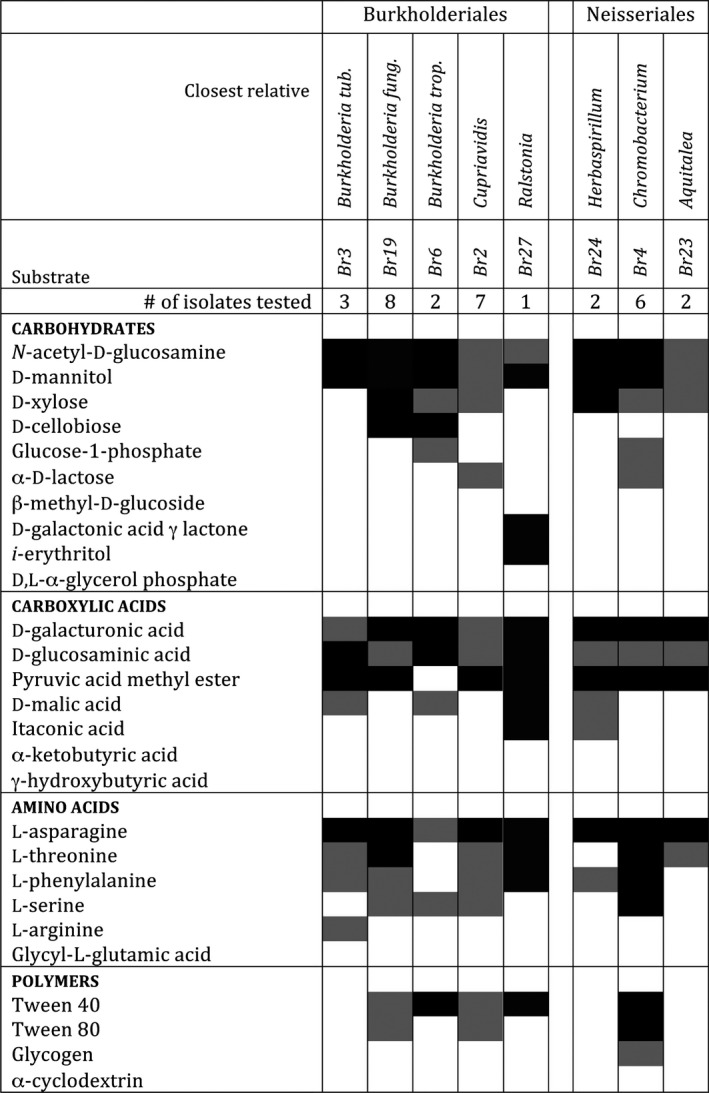
Metabolic profiling for bromeliad‐associated bacterial isolates using 31 defined substrates in six categories (carbohydrates, carboxylic acids, amino acids, polymers, amines, or phenols) with Biolog EcoPlates. Levels of metabolic utilization (as tetrazolium chloride reduction measured spectrophotometrically at 590 nm) were determined for 1–8 isolates of each type, as compared with a sterile water‐only substrate control. Use of amines or phenols was negative for all isolates and, thus, not shown. Black boxes indicate positive values and gray boxes indicate weakly positive values, or capabilities that were variable within isolate types. Empty boxes indicate no detectable activity.
Discussion
Diverse microorganisms have been recovered from tropical plant sources, including floral nectar, tree holes, Heliconia bracts, and bromeliad tank water (Walker et al. 1991; Goffredi et al. 2011a,b; Alvarez‐Pérez et al. 2012; Barbosa et al. 2012). While culture‐based methods are greatly limited in their assessment of the entire microbial community, these findings indicate the presence of a complex aquatic plant microbiota in the rainforest. Tank‐forming, epiphytic plants within the family Bromeliaceae are native to neotropical forests. They have unique foliar tanks that act as basins for rainwater storage, and are home to an array of microbes (Benzing 1970; Bermudes and Benzing 1991; Brighigna et al. 1992; Inselsbacher et al. 2007; Winkler and Zotz 2009; Goffredi et al. 2011a,b). The environment within a bromeliad tank can be acidic (as low as pH 3.5), with suboxic conditions (<0.5 ppm O2), and temperatures far above ambient (up to 44°C; Bernal Matarrita, pers comm). Bacterial residents must, therefore, adapt to these conditions in order to survive in this unusual environment. Indeed, most of the betaproteobacteria isolated in this study demonstrated tolerances for low pH and high temperature, as compared with close relatives (Tables 1, S1–S3), and were related to those known to tolerate, or even prefer, low‐oxygen conditions. Betaproteobacteria isolates belonged to heterotrophic groups that either are known to associate with plants or are found in tropical soils or freshwater habitats, including members of the orders Burkholderiales and Neisseriales. In a previous, molecular‐based survey of bacterial communities within Costa Rican bromeliad tanks, Burkholderiales and Neisseriales comprised ~2–7% of the total bacterial community recovered, based on 16S rRNA genes (Goffredi et al. 2011a).
Bromeliad‐associated Burkholderiales
The presence of the Burkholderiales has been previously observed for bromeliad tanks (Goffredi et al. 2011a). In this study, members of the genus Burkholderia (family Burkholderiaceae), in particular, were similarly recovered. Their common association with plants is illustrated by early descriptions of Burkholderia as pathogens of onions (at the time named Phytomonas spp; Burkholder 1950), yet since then, they have also been recovered from numerous environments, including acidic swamps and tropical soils (Vandamme et al. 2002; Valverde et al. 2003; Yang et al. 2006; Aizawa et al. 2010). The three Burkholderia‐related bacterial types isolated from the Costa Rican bromeliad tank cluster within the nonpathogenic “plant‐beneficial‐environmental” clade, and were most closely related to B. tropica, B. fungorum, and B. tuberum (Fig. 2; Coenye et al. 2001; Ussery et al. 2009; Suárez‐Moreno et al. 2012). These species are generally reported as beneficial for plants both casually and intimately (Reis et al. 2004; Barrett and Parker 2006; Kim et al. 2006; Omarjee et al. 2008; Suárez‐Moreno et al. 2012). Burkholderia are known for their versatile ability to degrade recalcitrant carbohydrates and aromatic compounds, such as l‐phenylalanine, and could thus contribute significantly to the turnover of plant‐derived organic carbon that collects in bromeliad tanks (Suárez‐Moreno et al. 2012). Carbon substrate utilization was versatile among the new, and described, Burkholderia species, suggesting effective utilization of numerous recalcitrant carbohydrates, including mannitol, cellobiose, xylose, and N‐acetyl‐d‐glucosamine (Tables 1, S1).
The sister genera Ralstonia and Cupriavidus (family Burkholderiaceae) were also recovered in bromeliad tank isolations. Both genera, subdivided nearly 10 years ago based on phenotypic differences, are inhabitants of soil, sludge, and wastewater or associated with plants (Coenye et al. 2003; Vandamme and Coenye 2004; Perez et al. 2008). For example, Cupriavidus taiwanensis, closely related to bromeliad isolate Br2, is known to be a dominant symbiont of the tropical legume Mimosa for which they provide fixed nitrogen, thereby increasing plant growth (Barrett and Parker 2006). Ralstonia species closely related to bromeliad isolate Br27 (ex. R. pickettii) have gained significant attention for their ability to degrade recalcitrant compounds (Ryan et al. 2007), and it is possible that similar isolates could contribute significantly to carbon turnover in bromeliad tanks. The Ralstonia‐related isolate from bromeliad tank water (Br27) was the only member of the genus capable of utilizing mannitol, while the Cupriavidus‐related isolate (Br2) was the only member of its genus capable of utilizing mannitol, xylose, and N‐acetyl‐d‐glucosamine, consistent with the exploitation of a habitat rich in plant‐based materials (Tables 1, S2).
Similarly, members of the genus Herbaspirillum (order Burkholderiales, family Oxalobacteraceae), which were recovered in two tank water isolation attempts, are primarily associated with the root surfaces and tissues of plants, for which they have been shown to promote growth (Baldani et al. 1986; Kirchhof et al. 2001; Valverde et al. 2003). As a genus, they are generally capable of utilizing plant‐sourced carbon substrates such as d‐galacturonic acid, mannitol, xylose, and N‐acetyl‐d‐glucosamine (Rothballer et al. 2006), as was the close relative isolated in this study, Br24 (Fig. 3).
Bromeliad‐associated Neisseriales
Bacteria within the genera Chromobacterium and Aquitalea (family Neisseriaceae) were isolated in this study at pH 6. Members of the genus are found in tropical and subtropical freshwater and terrestrial environments (Brazilian National Genome Project Consortium 2003; Martin et al. 2007; Young et al. 2008; Kampfer et al. 2009). The closest relative of the bromeliad‐associated isolate Br4, Chromobacterium subtsugae, has been recovered from soil rich in hemlock leaves and metabolizes plant compounds such as galacturonic acid and l‐phenylalanine (Martin et al. 2007). Species within this relatively new genus vary in their ability to live in anoxic environments (Leifson 1956). Well‐developed bromeliad tanks are highly stratified, with dramatically higher oxygen within 1 cm of the surface of the tank (up to 8 ppm O2), compared to <1 ppm O2 in the bottom (Goffredi et al. 2011b). It is, therefore, possible that Chromobacterium‐related members of the tank community inhabit the upper layers of the basin where not only higher O2 conditions persist, but also higher pH conditions (by as much as 1 pH unit; Goffredi et al. 2011b). Exclusively, bromeliad isolates related to Chromobacterium were recovered in this study at pH 6 and were unable to proliferate at pH 4. Nevertheless, isolate Br4 had a pH range shifted slightly lower, as well as the unique capability to metabolize d‐mannitol and xylose, compared to other members of Chromobacterium (Tables 1, S3). Members of the genus Aquitalea (family Neisseriaceae) are facultative anaerobes isolated previously from similarly organic‐rich environments, such as humic lakes and wetland peat (Lau et al. 2006; Lee et al. 2009). They are distinct from Chromobacterium in their inability to use l‐phenylalanine and their reduced tolerance to higher salinity and temperatures. The Aquitalea‐related isolate Br23 from this study uniquely displayed the ability to metabolize starch, d‐galacturonic acid, mannitol, xylose, and n‐acetyl‐d‐glucosamine (Tables 1, S3).
Community function: breakdown of organic substrates
Bromeliads, with their unique ∨‐shaped central rosette and trough‐like leaves, are known to collect large amounts of both plant‐ and animal‐derived material (Benzing et al. 1972; Pittl et al. 2010). As a consequence, these catchments are an ideal site for microbial decomposition. The ability of all bacterial isolates examined in this study to utilize plant‐derived compounds indicates their likely facilitation in the turnover of leaves and plant debris entrained within the tanks from the overlying rainforest canopy. For example, all isolate types could use d‐galacturonic acid, a component of the plant cell wall (Isherwood et al. 1953) and mannitol, a sugar alcohol produced by plants during times of osmotic stress to balance inner solute concentrations (Stoop et al. 1996). The ability to use mannitol was confirmed using APIZYM analysis and growth in phenol red fermentation agar (data not shown). Similarly, six isolate types were capable of using d‐xylose, a key component of pectin (Mohnen 2008) and three isolate types used l‐phenylalanine, which is synthesized by chloroplasts, and serves as a precursor to many plant‐derived metabolites (Jung, 1986). The ability to use these substrates was unique, in some cases, compared to close relatives (Table 1), and may prove to be an important component for the niche specialization of bacteria within the bromeliad tank.
A further capability demonstrated by all eight isolates was the use of N‐acetyl‐d‐glucosamine, a monomeric unit of chitin. While not a unique trait, these bacteria do appear to be poised to also aid in the breakdown of chitinous remains of insect exoskeletons, and fungal cell walls, prevalent within the bromeliad tank environment. Chitinases have been previously recovered from bromeliad tanks, including one from Chromobacterium (Goffredi et al. 2011a), confirming the likely role of microbial residents in the biological remineralization of recalcitrant chitin in tropical forests. The ability to use N‐acetyl‐d‐glucosamine and/or chitin directly was confirmed using APIZYM analysis and growth in phenol red fermentation agar (data not shown).
It is important to note that fungi are also likely to play an important role in the hydrolysis of plant material trapped in bromeliad tanks. In a previous metatranscriptomic analysis, genes from eight fungal classes, including Ascomycota and Basidiomycota, were recovered from the water catchments formed by Costa Rican bromeliads (Goffredi et al. 2015). It will therefore be important, in the future, to examine the likely involvement of fungi in the stability and functioning of bromeliad ecosystems.
Conclusion
Betaproteobacteria from two orders were specifically isolated from the unique aquatic habitat within a bromeliad tank. Compared to close relatives, the isolates displayed high temperature and comparatively low pH optima, reflecting the tropical, acidic nature of bromeliad tanks. Metabolic activity profiles were aligned with a role in the breakdown of plant‐sourced material. Further, these activities overlapped between the Burkholderiales, isolated at pH 4, and the Neisseriales, isolated at pH 6, suggesting that plant material decomposition occurs in tanks of both pH extremes. Despite their importance within neotropical habitats, the specific nutritional strategy used by bromeliads remains a mystery. We suggest that future efforts focus on expanding current knowledge on the specific role of bacterial, as well as fungal, mineral leeching, nitrogen fixation, and metabolic exchanges with these important ecosystem‐structuring plants.
Conflict of Interest
The authors declare no conflict of interest in relation to the submitted work.
Supporting information
Table S1. Phenotypic characteristics of bromeliad tank strains in comparison to other closely related members of the genus Burkholderia.
Table S2. Phenotypic characteristics of bromeliad tank strains in comparison to other members of the genera Ralstonia and Cupriavidus.
Table S3. Phenotypic characteristics of bromeliad tank strains in comparison to other members of the genera Chromobacterium and Aquitalea.
Acknowledgments
The authors thank Dr. Gretchen North and Dr. Beth Braker (Occidental College) for scientific and logistical support, as well as mentorship of students; Bernal Matarrita for his laboratory support at La Selva; Dr. Deedra McClearn and Dr. Carlos de la Rosa and members of the Organization for Tropical Studies for their support of this project; Occidental College undergraduates involved in the Spring 2012 semester of Microbial Diversity (Bio325). Funding for this project included, in part, a NSF grant to B. Braker (Occidental College, OISE‐0526551), as well as the Undergraduate Research Center (Academic Student and Summer Research Projects), and a Faculty Enrichment grant from Occidental College to S.K.G.
MicrobiologyOpen 2016; 5(3): 479–489
References
- Aizawa, T. , Nguyen B. V., Nakajima M., and Sunairi M.. 2010. Burkholderia heleia sp. nov., a nitrogen‐fixing bacterium isolated from an aquatic plant, Eleocharis dulcis, that grows in highly acidic swamps in actual acid sulfate soil areas of Vietnam. Int. J. Syst. Evol. Microbiol. 60:1152–1157. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F. , Madden T. L., Schäffer A. A., Zhang J., Z. Zhang , Miller W., et al. 1997. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Pérez, S. , Herrera C. M., and de Vega C.. 2012. Zooming‐in on floral nectar: a first exploration of nectar‐associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 80:591–602. [DOI] [PubMed] [Google Scholar]
- Baldani, J. I. , Baldani V. L. D., Seldin L., and J. Dobereiner . 1986. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root‐associated nitrogen‐fixing bacterium. Int. J. Syst. Bacteriol. 30:86–93. [Google Scholar]
- Barbosa, A. C. , Morais C. G., Morais P. B., Rosa L. H., Pimenta R. S., Lachance M. A., et al. 2012. Wickerhamiella pagnoccae sp. nov. and Candida tocantinsensis sp. nov., two ascomycetous yeasts from flower bracts of Heliconia psittacorum (Heliconiaceae). Int. J. Syst. Evol. Microbiol. 62:459–464. [DOI] [PubMed] [Google Scholar]
- Barrett, C. F. , and Parker M. A.. 2006. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 72:1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing, D. H. 1970. Availability of exogenously supplied nitrogen to seedling of Bromeliaceae. Bull. Torrey Bot. Club 97:154–159. [Google Scholar]
- Benzing, D. H. 2000. Bromeliaceae: profile of an adaptive radiation. Cambridge University Press, Cambridge. [Google Scholar]
- Benzing, D. H. , Derr J. A., and Titus J. E.. 1972. The water chemistry of microcosms associated with the bromeliad Aechmea bracteata . Am. Midl. Nat. 87:60–70. [Google Scholar]
- Bermudes, D. , and Benzing D. H.. 1991. Nitrogen fixation in association with Ecuadorean bromeliads. J. Trop. Ecol. 7:531–536. [Google Scholar]
- Brazilian National Genome Project Consortium . 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. PNAS 100:11660–11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brighigna, L. , Montaini P., Favilli F., and Trejo A. C.. 1992. Role of nitrogen‐fixing bacterial microflora in the epiphytism of Tillandsia (Bromeliaceae). Am. J. Bot. 79:723–727. [Google Scholar]
- Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115–117. [Google Scholar]
- Chen, W.‐M. , James E. K., Coenye T., Chou J. H., E. Barrios , de Faria S. M., Elliott G. N., Sheu S.‐Y., Sprent J. I., and Vandamme P.. 2006. Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int J Syst Evol Microbiol 56:1847–1851. [DOI] [PubMed] [Google Scholar]
- Chen, W.‐M. , Laevens S., Lee T.‐M., Coenye T., De Vos P., Mergeay M., et al. 2001. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51:1729–1735. [DOI] [PubMed] [Google Scholar]
- Clarke, K. R. , and Gorley R. N.. 2006. PRIMER v6: user Manual/Tutorial. PRIMER‐E, Plymouth, UK. [Google Scholar]
- Coenye, T. , Laeven S., Willems A., Ohlen M., Hannant W., J. R. Govan , et al. 2001. Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol. 51:1099–1107. [DOI] [PubMed] [Google Scholar]
- Coenye, T. , Goris J., De Vos P., Vandamme P., and LiPuma J. J.. 2003. Classification of Ralstonia pickettii‐like isolates from the environment and various clinical samples as Ralstonia insidiosa sp. nov. Int. J. Syst. Evol. Microbiol. 53:1075–1080. [DOI] [PubMed] [Google Scholar]
- De Baere, T. , Steyaert S., Wauters G., De Vos P., Goris J., Coenye T., et al. 2001. Classification of Ralstonia pickettii biovar 3/’thomasii’ strains (Pickett 1994) and of new isolates related to nosocomial recurrent meningitis as Ralstonia mannitolytica sp. nov. Int. J. Syst. Evol. Microbiol. 51:547–558. [DOI] [PubMed] [Google Scholar]
- Dedysh, S. N. , Pankratov T. A., Belova S. E., Kulichevskaya I. S., and Liesack W.. 2006. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 27:2110–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, D. 1983. Phytotelmata: terrestrial plants as hosts for aquatic insect communities Pp. 1–28 in Frank J. H. and Lounibos L. P., eds. Phytotelmata: flora and fauna. Plexus, New Jersey. [Google Scholar]
- Goffredi, S. K. , Jang G., Woodside W. T., and Ussler W. III. 2011a. Bromeliad catchments as habitats for methanogenesis in tropical rainforest canopies. Front. Microbiol. 2:256. doi:10.3389/fmicb.2011.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffredi, S. K. , Kantor A. H., and Woodside W. T.. 2011b. Aquatic microbial habitats within a neotropical rainforest: bromeliads and pH‐associated trends in bacterial diversity and compostion. Microb. Ecol. 61:529–542. [DOI] [PubMed] [Google Scholar]
- Goffredi, S. K. , Jang G., and Haroom M.. 2015. Transcriptomics in the tropics: total RNA‐based profiling of Costa Rican bromeliad‐associated communities. Comput. Struct. Biotechnol. J. 13:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes‐Souza, B. A. , Mendes G. B., Bento L., Morotta H., Santorao A. L., Esteves F. A., et al. 2006. Limnological parameters in the water accumulated in tropical bromeliads. Acta Limnol. Bras. 18:47–53. [Google Scholar]
- Inselsbacher, E. , Cambui C. A., Richter A., Stange C. F., H. Mercier , and Wanek W.. 2007. Microbial activities and foliar uptake of nitrogen in the epiphytic bromeliad Vriesea gigantea . New Phytol. 175:311–320. [DOI] [PubMed] [Google Scholar]
- Isherwood, F. A. , Chen Y. T., and Mapson L. W.. 1953. Synthesis of L‐ascorbic acid in plants and animals. Biochemisty 56:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, E. , Zamir L. O., and Jensen R. A.. 1986. Chloroplasts of higher plants synthesize l‐phenylalanine via l‐arogenate. Proc Natl Acad Sci USA 83:7231–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfer, P. , Busse H., and Scholz H.. 2009. Chromobacterium piscinae sp. nov. and Chromobacterium pseudoviolaceum sp. nov., from environmental samples. Int. J. Syst. Evol. Microbiol. 59:2486–2490. [DOI] [PubMed] [Google Scholar]
- Kim, H.‐B. , Park M.‐J., Yang H.‐C., An D.‐S., Jin H.‐Z., and Yang D.‐C.. 2006. Burkholderia ginsengisoli sp. nov., a beta‐glucosidase producing bacterium isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 56:2529–2533. [DOI] [PubMed] [Google Scholar]
- Kirchhof, G. , Eckert B., Stoffels M., Baldani J. I., Reis V. M., and Hartmann A.. 2001. Herbaspirillum frisingense sp. nov., a new nitrogen‐fixing bacterial species that occurs in C4‐fibre plants. Int. J. Syst. Evol. Microbiol. 51:157–168. [DOI] [PubMed] [Google Scholar]
- Laessle, A. M. 1961. A micro‐limnological study of Jamaican bromeliads. Ecology 42:499–517. [Google Scholar]
- Lane, D. J. 1991. 16S/23S rRNA sequencing Pp. 115–175 in Stackebrandt E. and Goodfellow M., eds. Nucleic acid techniques in bacterial systematics. Wiley, New York. [Google Scholar]
- Lau, H. T. , Faryna J., and Triplett E. W.. 2006. Aquitalea magnusonii gen. nov., sp. nov., a novel Gram‐negative bacterium isolated from a humic lake. Int. J. Syst. Evol. Microbiol. 56:867–871. [DOI] [PubMed] [Google Scholar]
- Lee, C. , Weon H., Kim Y., Son J., Yoon S., Koo B., et al. 2009. Aquitalea denitrificans sp. nov., isolated from a Korean wetland. Int. J. Syst.Evol. Microbiol. 59:1045–1048. [DOI] [PubMed] [Google Scholar]
- Leifson, E. 1956. Morphological and physiological characteristics of the genus Chromobacterium . J. Bacteriol. 71:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkar, N. S. , and Casida L. E. Jr. 1987. Cupriavidus necator gen. nov., sp. nov.: a nonobligate bacterial predator of bacteria in soil. Int. J. Syst. Bacteriol. 37:323–326. [Google Scholar]
- Martin, P. A. W. , Gundersen‐Rindal D., Blackburn M., and J. Buyer . 2007. Chromobacterium substsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int. J. Syst. Evol. Microbiol. 57:993–999. [DOI] [PubMed] [Google Scholar]
- Mohnen, D. 2008. Pectin structure and synthesis. Curr. Opin. Plant Biology 11:266–277. [DOI] [PubMed] [Google Scholar]
- Omarjee, J. , Balandreau J., Spaull V. W., and Cadet P.. 2008. Relationships between Burkholderia populations and plant parasitic nematodes in sugarcane. Appl. Soil Ecol. 391:1–14. [Google Scholar]
- Perez, S. A. , Mejia L., Fegan M., and Allen C.. 2008. Diversity and distribution of Ralstonia solanacearum strains in Guatemala and rare occurrence of tomato fruit infection. Plant. Pathol. 57:320–331. [Google Scholar]
- Pittl, E. , Innerebner G., Wanek W., and Insam H.. 2010. Microbial communities of arboreal and ground soils in the Esquinas rainforest, Costa Rica. Plant Soil 329:65–74. [Google Scholar]
- Reis, V. M. , Estrada‐ de los Santos P., Tenorio‐Salgado S., Vogel J., Stoffels M., Guyon S., et al. 2004. Burkholderia tropica sp. nov., a novel nitrogen‐fixing, plant‐associated bacterium. Int. J. Syst. Evol. Microbiol.. 54: 2155–2162. [DOI] [PubMed] [Google Scholar]
- Richardson, B. A. 1999. The bromeliad microcosm and the assessment of faunal diversity in a neotropical forest. Biotropica 31:65–74. [Google Scholar]
- Rothballer, M. , Schmid M., Klein I., Gattinger A., Grundmann S., and Hartmann A.. 2006. Herbaspirillum hiltneri sp. nov., isolated from surface sterilized wheat roots. Int. J. Syst. Evol. Microbiol. 56:1341–1348. [DOI] [PubMed] [Google Scholar]
- Rui, J. , Peng J., and Lu Y.. 2009. Succession of bacterial populations during plant residue decomposition in rice field soil. Appl. Environ. Microbiol. 75:4879–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, M. P. , Pembroke J. T., and Adley C. C.. 2007. Ralstonia pickettii in environmental biotechnology: potential and applications. J. Appl. Microbiol. 103:754–764. [DOI] [PubMed] [Google Scholar]
- Sessitsch, A. , Coenye T., Sturz A. V., Vandamme P., Barka E. A., Salles J. F., et al. 2005. Burkholderia phytofirmans sp. nov., a novel plant‐associated bacterium with plant‐beneficial properties. Int. J. Syst. Evol. Microbiol. 55:1187–1192. [DOI] [PubMed] [Google Scholar]
- Stoop, J. M. H. , Williamson J. D., and Pharr D. M.. 1996. Mannitol metabolism in plant: a method for coping with stress. Trends Plant Sci. 1:139–144. [Google Scholar]
- Suárez‐Moreno, R. Z. , Caballero‐Mellado J., Coutinho B. G., Mendonça‐Previato L., James E. K., and Venturi V.. 2012. Common features of environmental and potentially beneficial plant‐associated Burkholderia . Microb. Ecol. 63:249–266. [DOI] [PubMed] [Google Scholar]
- Sugden, A. M. , and Robins R. J.. 1979. Aspects of the ecology of vascular epiphytes in colombian cloud forests, I. The distribution of the epiphytic flora. Biotropica 11:173–188. [Google Scholar]
- Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland. [Google Scholar]
- Ussery, D. W. , Kiil K., Lagesen K., Sicheritz‐Ponten T., J. Bohlin , and Wassenaar T. M.. 2009. The genus Burkholderia: analysis of 56 genomic sequences. Genome Dyn. 6:140–157. [DOI] [PubMed] [Google Scholar]
- Valverde, A. , Velazquez E., Gutierrez C., Cervantes E., A. Ventosa , and Igual J. M.. 2003. Herbaspirillum lusitanum sp. nov., a novel nitrogen‐fixing bacterium associated with root nodules of Phaseolus vulgaris . Int. J. Syst. Evol. Microbiol. 53:1979–1983. [DOI] [PubMed] [Google Scholar]
- Vandamme, P. , and Coenye T.. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54:2285–2289. [DOI] [PubMed] [Google Scholar]
- Vandamme, P. , Goris J., Coenye T., Hoste B., Janssens D., Kersters K., De Vos P., and Falsen E.. 1999. Assignment of Centers for Disease Control group IVc‐2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int J Syst Bacteriol 49:663–669. [DOI] [PubMed] [Google Scholar]
- Vandamme, P. , Goris J., Chen W. M., de Vos P., and A. Willems . 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507–512. [DOI] [PubMed] [Google Scholar]
- Vaneechoutte, M. , Kampfer P., De Baere T., Falsen E., and Verschraegen G.. 2004. Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. Int. J. Syst. Evol. Microbiol. 54: 317–327. [DOI] [PubMed] [Google Scholar]
- Walker, E. D. , Lawson D. L., Merritt R. W., Morgan W. T., and Klug M. J.. 1991. Nutrient dynamics, bacterial populations, and mosquito productivity in tree hole ecosystems and microcosms. Ecology 72:1529–1546. [Google Scholar]
- Winkler, U. , and Zotz G.. 2009. Highly efficient uptake of phosphorus in epiphytic bromeliads. Ann. Bot. 103:447–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. C. , Im W. T., Kim K. K., An D. S., and Lee S. T.. 2006. Burkholderia terrae sp. nov., isolated from a forest soil. Int. J. Syst. Evol. Microbiol. 56:453–457. [DOI] [PubMed] [Google Scholar]
- Yarwood, S. A. , Myrold D. D., and Högberg M. N.. 2009. Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microbiol. Ecol. 70:151–162. [DOI] [PubMed] [Google Scholar]
- Young, C. , Arun A. B., Lai W., Chen W., Chao J., Shen F., et al. 2008. Chromobacterium aquaticum sp. nov., isolated from spring water samples. Int. J. Syst. Evol. Microbiol. 58:877–880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Phenotypic characteristics of bromeliad tank strains in comparison to other closely related members of the genus Burkholderia.
Table S2. Phenotypic characteristics of bromeliad tank strains in comparison to other members of the genera Ralstonia and Cupriavidus.
Table S3. Phenotypic characteristics of bromeliad tank strains in comparison to other members of the genera Chromobacterium and Aquitalea.


