Figure 1.
Comparison of ComP Orthologs in N. meningitidis and N. subflava, and of Their Cognate DUS Variants
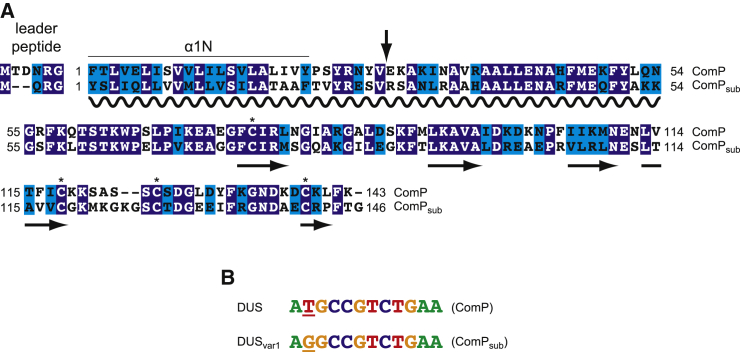
(A) Sequence alignment of ComP and ComPsub from N. meningitidis 8013 and N. subflava NJ9703, produced using Clustal Omega. Amino acids are shaded in dark blue (when identical) or light blue (when highly similar), or non-shaded (when non-conserved). Relevant structural and functional features are highlighted. The proteins start with a conserved N-terminal sequence motif that defines all type IV pilins, the class III signal peptide (Szabó et al., 2007). This motif consists of a hydrophilic leader peptide, which is cleaved by the pre-pilin peptidase PilD, followed by a stretch of 21 predominantly hydrophobic residues that forms an extended α helix, which is the main assembly interface of subunits within filaments (Berry and Pelicic, 2015). To facilitate purification, we produced the recombinant proteins without their 28 N-terminal residues, depicted by an arrow. The four Cys residues that form two crucial disulfide bonds are identified by asterisks. The soluble portions that have been purified in this study, as well as the different structural motifs, are also highlighted.
(B) Sequence alignment of DUS and DUSvar1 found in N. meningitidis and N. subflava genomes, respectively. These 12-bp motifs (Ambur et al., 2007) differ by just one base, which is underlined.