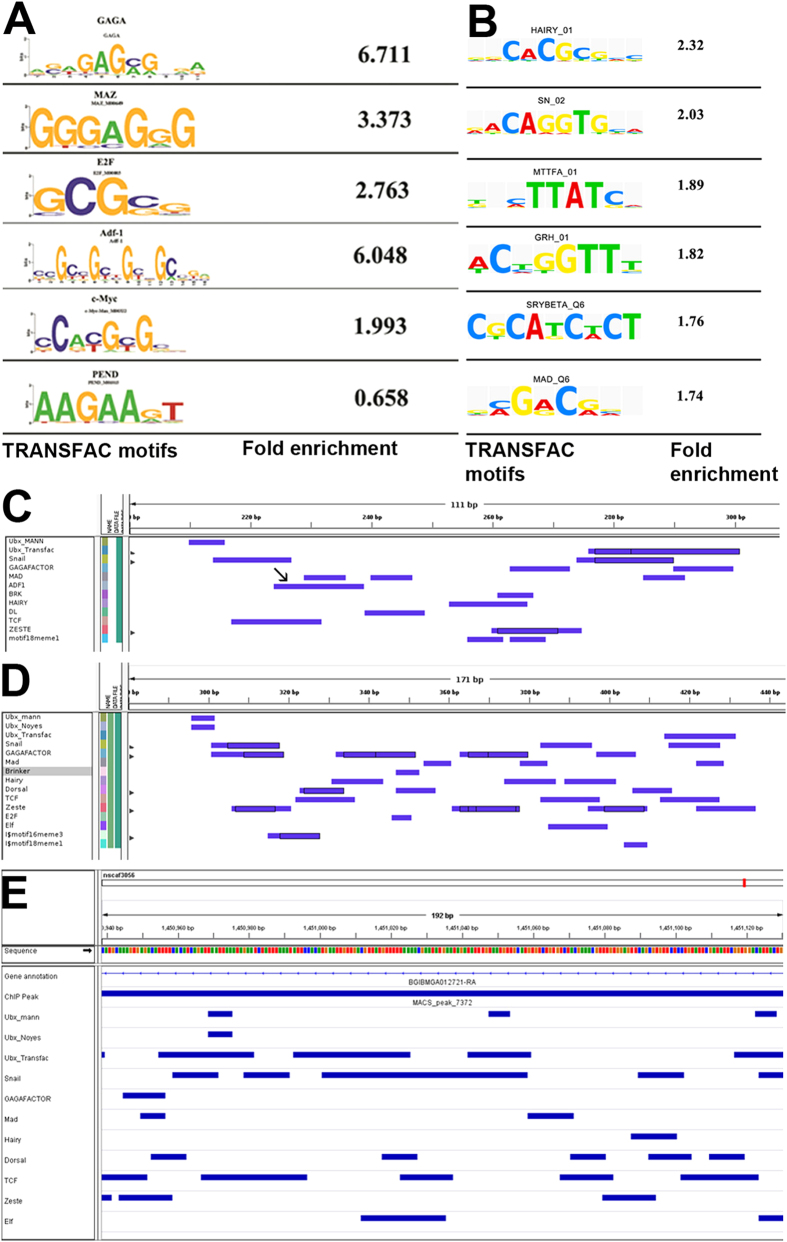
Figure 6. Bioinformatics analyses of putative enhancer sequences of targets of Ubx in Apis and Bombyx.
(A,B) Enrichment of binding sites for various TFs in the sequences pulled down by anti-UbxApis from Apis (A) and by anti-UbxBombyx from Bombyx (B) hindwing buds identified using TRANSFAC. Motifs are based on information available for TFs available in Drosophila. These are similar to the ones reported for putative targets of Ubx in Drosophila7. In both (A) and (B), numbers indicate fold differences in the frequency of occurrence of a given motif between sequences pulled down by ChIP and non-coding sequences randomly collated from Apis or Bombyx genome. Similar enrichment for binding sites for GAF, Adf-1, E2F etc. is reported for Drosophila7. (C–E) TRANSFAC analysis of quadrant enhancer of vg in Drosophila (C) to its equivalent region in vg gene of Apis (D) and Bombyx (E). Note a similar array of TFs bind around Ubx binding sites in all the three species. However, Adf-1 binding site is absence in Apis and Bombyx.