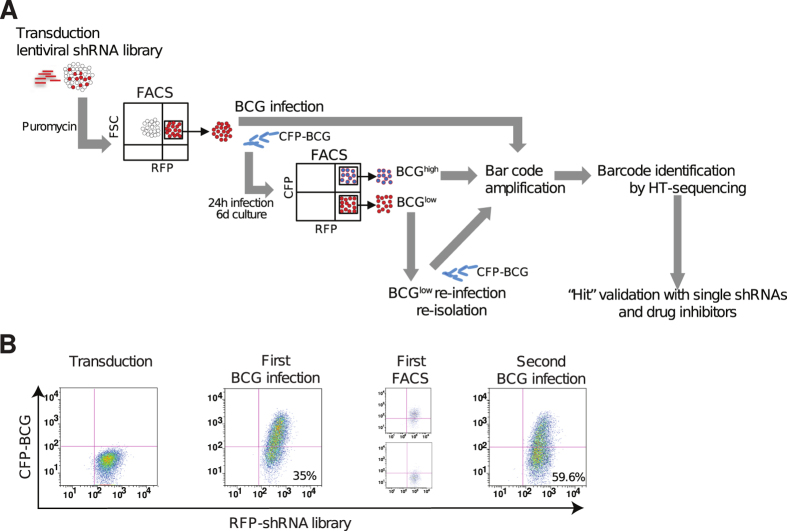
Figure 1. Experimental approach for screening the DECIPHER™ pooled shRNA library in mycobacteria-infected THP1 cells.
(A) THP1 cells were transduced with lentiviral particles encoding 27,500 shRNA sequences targeting 5,043 genes (5–6 shRNAs/mRNA) and expressing red fluorescence protein (RFP). The shRNA+ clones were then selected (puromycin, RFP expression), expanded in culture and infected with BCG expressing cyan fluorescent protein (CFP-BCG). Six days after infection, cell clones with high bacterial loads (BCGhigh) and those that were either uninfected or harbor low number of bacteria (BCGlow) were separated by FACS based on levels of CFP expression. After expansion in culture, BCGlow cells were re-infected and re-isolated to enrich the phenotype. DNA was isolated from BCGhigh-, BCGlow- and uninfected cells, and barcodes were PCR-amplified. Barcodes sequences were identified and quantified by high-throughput Illumina sequencing (HT-sequencing). “Hits” were validated in THP1 cells and MDM with single shRNAs and small molecule inhibitors. (B) THP1 cells were analyzed by flow cytometry after transduction with lentiviral particles and selection with puromycin and FACS (Transduction), first round of infection with BCG (First BCG infection), purification by FACS of BCGhigh and BCGlow cells after first infection (First FACS) and re-infection of BCGlow cells (Second BCG infection).