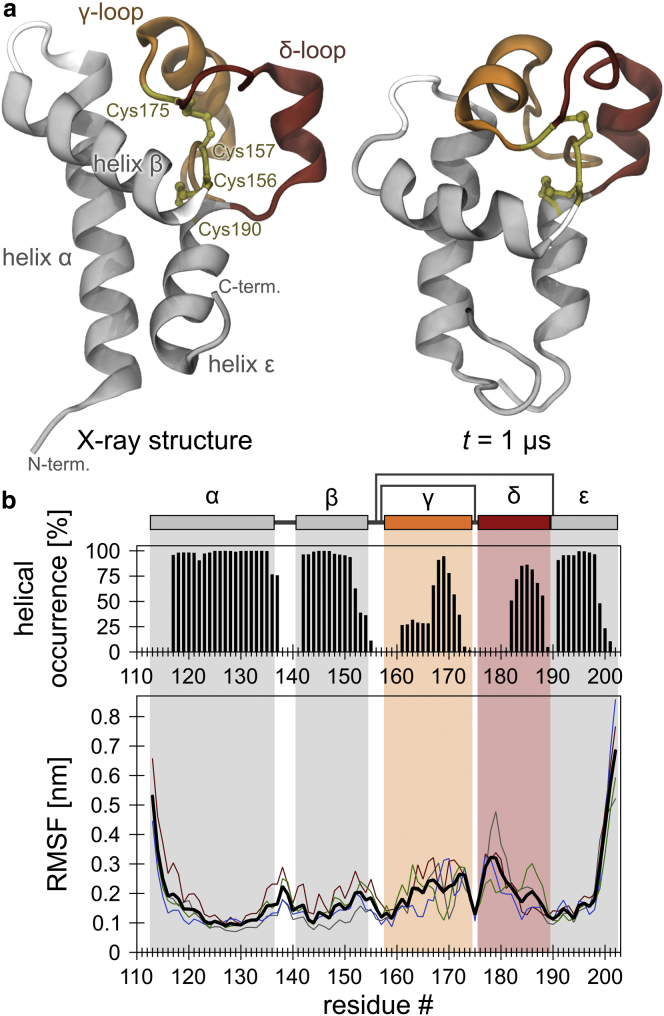
Figure 1.
Helical stability and backbone flexibility of the isolated LEL of CD81. (a) Left: CD81LEL initial conformation (chain A of the x-ray structure (20) (PDB: 1G8Q)). Right: after a 1 μs unbiased, atomistic MD simulation (run B; see also Fig. S1b). (b) Top: occurrence of helical structure per residue (based on the ϕ/ψ dihedral angles) over time, averaged over four runs. Bottom: Cα RMSF as a measure of the local protein backbone flexibility. The highest flexibility of the fluctuating termini is due to the lack of anchoring to the TM segments present in the full-length protein (compare with Fig. 2b, which shows the flexibility of this region in the full-length protein). Colored lines show the flexibility profiles from the individual MD runs; average (n = 4) is shown in black. The pictogram shows the CD81 LEL domain structure, including cysteine disulfide bridges indicated by additional black lines connecting residues 157 with 175, and 156 with 190. To see this figure in color, go online.