Figure 1.
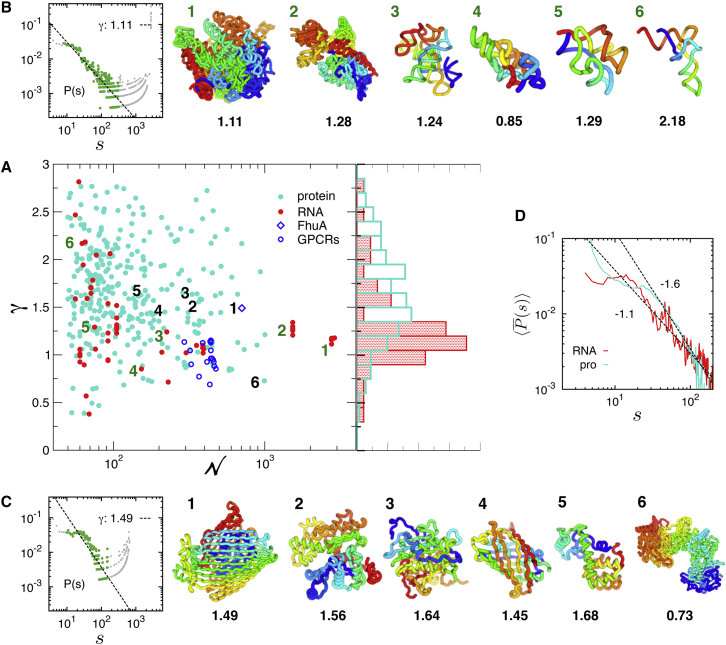
Contact probability scaling exponent, γ, and chain configurations of RNA and proteins. (A) The value γ versus N obtained for 60 individual RNA (data in red) and 324 proteins (data in cyan), whose γ-value is obtained from the power-law fit to P(s) with the correlation coefficient (c.c.) > 0.9 (see Fig. S4 for γ versus N plot with error bars, and Tables S1 and S2 in the Supporting Material for PDB entries used here). The data points for FhuA and GPCRs are included for further discussion, although c.c.. Histograms of γ, p(γ) for RNA (γ ≈ 1.30 ± 0.44) and proteins (γ = 1.65 ± 0.50) are shown on the right. (B) Representative structures of RNA in the rainbow coloring scheme from 5′ (blue) to 3′ (red), indexed with the number in γ versus N plot. Depicted are the structures of 1) a large subunit of rRNA (PDB: 2O45 (66), γ = 1.11); 2) a small subunit of rRNA (PDB: 2YKR, γ = 1.28) (67); 3) Twort group I ribozyme (PDB: 1Y0Q, γ = 1.24) (68); 4) A-type ribonuclease P (PDB: 1U9S, γ = 0.85) (69); 5) TPP-riboswitch (PDB: 3D2G, γ = 1.29) (70); and 6) tRNA (PDB: 1VTQ, γ = 2.18). P(s), which provides γ-value, is shown for a large subunit of rRNA on the left corner. The scaling exponent (γ) of is obtained from the fit (dashed line) to the data points in green; the data in gray are excluded from the fit (see the Supporting Material for details of fitting procedure). (C) Protein structures in the rainbow coloring scheme from the N- (blue) to the C-terminus (red). Depicted in (C) are the structures of 1) FhuA (PDB: 1QJQ, γ = 1.49) (71); 2) an actin monomer (PDB: 1J6Z, γ = 1.56) (72); 3) metacaspase (PDB: 4AF8, γ = 1.64) (73); 4) green fluorescent protein (PDB: 1EMA, γ = 1.45) (74); 5) T4 lysozyme (PDB: 2LZM, γ = 1.68) (32, 75); and 6) Chondroitin Sulfate ABC lyase I (PDB: 1HN0, γ = 0.73) (25). P(s) for FhuA is shown on the left corner. (D) The mean contact probabilities, , calculated over the RNA and protein structures in the PDB. To see this figure in color, go online.