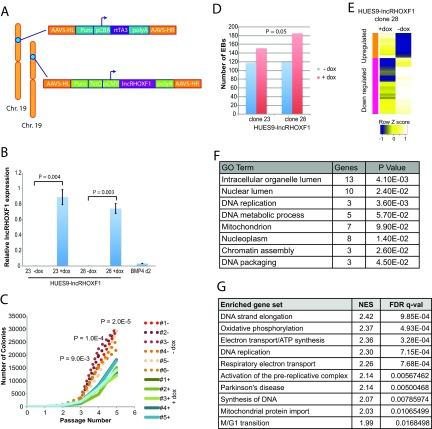
FIG 3.
Overexpression of lncRHOXF1 in undifferentiated hESCs promotes cellular differentiation by changing gene expression. (A) Targeting constructs for inducible expression of lncRHOXF1 RNA from the AAVS locus on chromosome 19 in hESCs. The reverse tetracycline transactivator (rtTA3) was constitutively expressed using the chicken β-actin (CBA) promoter. (B) qRT-PCR analysis of lncRHOXF1 expression in uninduced (−dox) and induced (+dox) hESCs, and BMP4/A/P-differentiated hESCs (for comparison). P values were determined using a two-tailed t test to compare untreated versus induced samples. Error bars denote standard deviations from the mean. (C) Overexpression of lncRHOXF1 RNA (clone 28) reduces cell growth, which was quantified based on the number of undifferentiated colonies during routine passaging. Five and six independent replicates for each condition (with or without doxycycline) were followed for 5 passages. Statistical significance was determined using a two-tailed paired t test comparing –dox and +dox samples. (D) Overexpression of lncRHOXF1 (clones 23 and 28) increases the number of embryoid bodies during differentiation. The P value was determined using a one-tailed t test to compare EB quantities between clones 23 and 28. (E) Microarray analyses of lncRHOXF1 overexpression in hESCs. The heat map indicates clustering of upregulated and downregulated genes for the 162 differentially expressed genes in HUES9-lncRHOXF1 cells (clone 28) treated or not with doxycycline. Biological replicates (triplicates for each sample) were averaged together to determine gene expression differences with lncRHOXF1 induction. (F) GO analysis for the 162 differentially expressed genes (red, upregulated; blue, downregulated) when lncRHOXF1 is overexpressed (+dox). P values were determined by using the DAVID online analysis tool. (G) GSEA of gene sets enriched in DOX-treated HUES9-lncRHOXF1 cells. NES, normalized enrichment score; FDR, false-discovery rate. FDR q values were calculated by using the GSEA tool.