FIG 2.
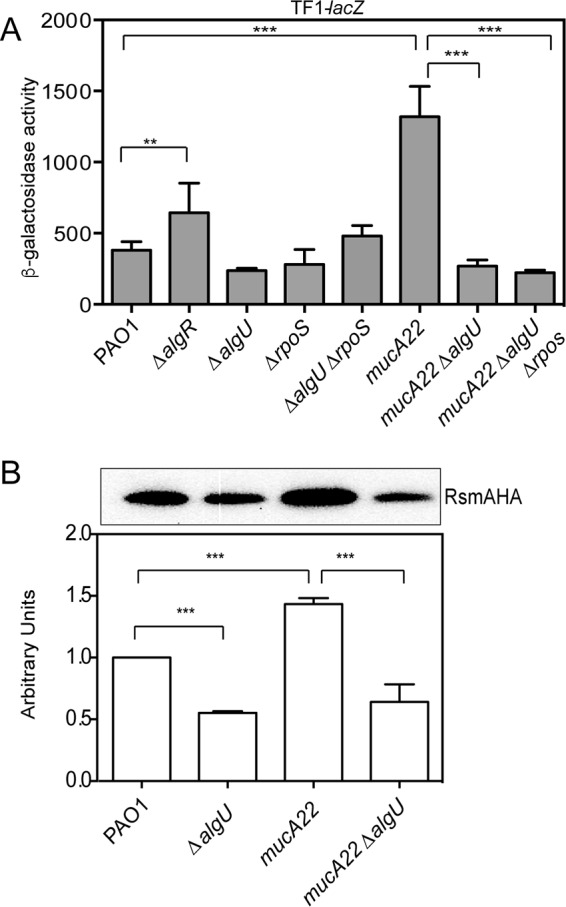
AlgU activates rsmA expression in mucA mutants. (A) Analysis of TF1-lacZ in mutant strains at 8 h of growth in LB broth. The TF1-lacZ transcriptional fusion was inserted into the attB site in the chromosomes of the indicated strains. Transcriptional fusion values are indicated as β-galactosidase activity. The values are the result of subtraction of the value for PAO1 containing the vector (miniCTXlacZ) alone (28 Miller units) from all test values. Fusion assays were all conducted at least three times. Determination of all significant differences from the results for the wild type was performed using a one-way analysis of variance and Tukey's posttest. **, P = 0.01; ***, P < 0.001. (B) (Top) Western analysis of RsmA in algU mutant strains. An rsmA-HA allele was constructed and introduced into various strains via homologous recombination, the strains were grown for 8 h, and whole-cell lysates were probed with anti-HA antibody. A single band was detected in all Western blots using the strains containing the rsmA-HA allele. A representative Western blot is shown. Western blot analysis was completed at least three times. The strains assayed are listed below the bottom panel. PAO1 without the rsmA-HA allele was run as a negative control (data not shown). (Bottom) Densitometry analysis was performed for each strain. Densitometry values, determined using ImageJ software, were standardized by dividing the intensity of the band in the Western blot by the intensity of the total protein in each lane stained with Coomassie. The result for PAO1 was set equal to 1, and the values for the other strains were normalized using the value for PAO1 as a reference. A one-way ANOVA with Tukey's posttest was used to determine statistical significance. ***, P < 0.0001.
