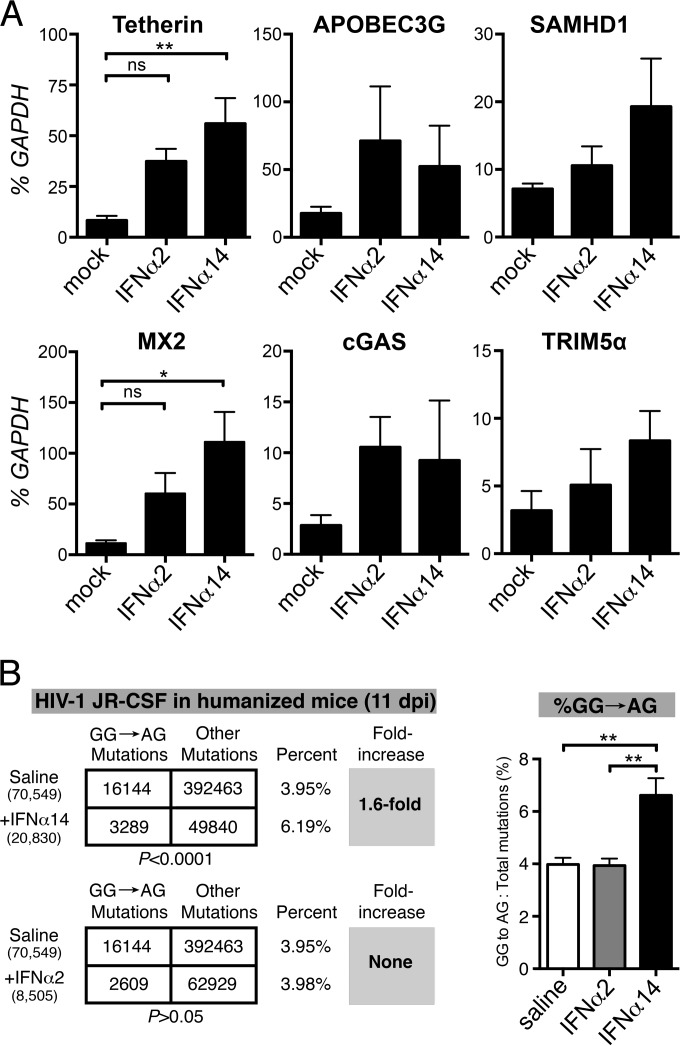
FIG 7.
Induction of ISGs. (A) The transcription levels of six ISGs encoding HIV-1 sensors or restriction factors were determined in splenocytes harvested from HIV-1-negative TKO-BLT mice 6 h after intravenous injection with a single 1.5 × 105 U dose of the indicated IFN or mock saline control injection. RNA levels were determined by qPCR and are expressed as a percentage (means + SEM) of the human housekeeping GAPDH transcripts detected in the same sample. n = 5 mice per group. Statistical analyses were done by one-way ANOVA with Tukey's posttest. ns, not significant; **, P < 0.01; *, P < 0.05. (B) Evaluation of APOBEC3G signature mutations in IFN-α-treated mice. A 350-bp segment of the HIV-1 gp41/nef region was amplified from lymph node DNA from humanized mice treated with saline (n = 5), IFN-α14 (n = 4), and IFN-α2 (n = 3) at 11 dpi for next-generation sequencing. (Left) DNA sequences from all mice per cohort were pooled, and the relative numbers of GG→AG mutations relative to the total number of mutations were compared using a 2-by-2 contingency test with Yates' correction. The numbers of sequences analyzed are shown in parentheses, and the percentages of GG→AG mutations relative to the total number of mutations are shown. (Right) Percentages (means + SD) of GG→AG mutations relative to the total number of mutations computed per mouse. **, P < 0.01 (one-way ANOVA with Bonferroni's posttest).