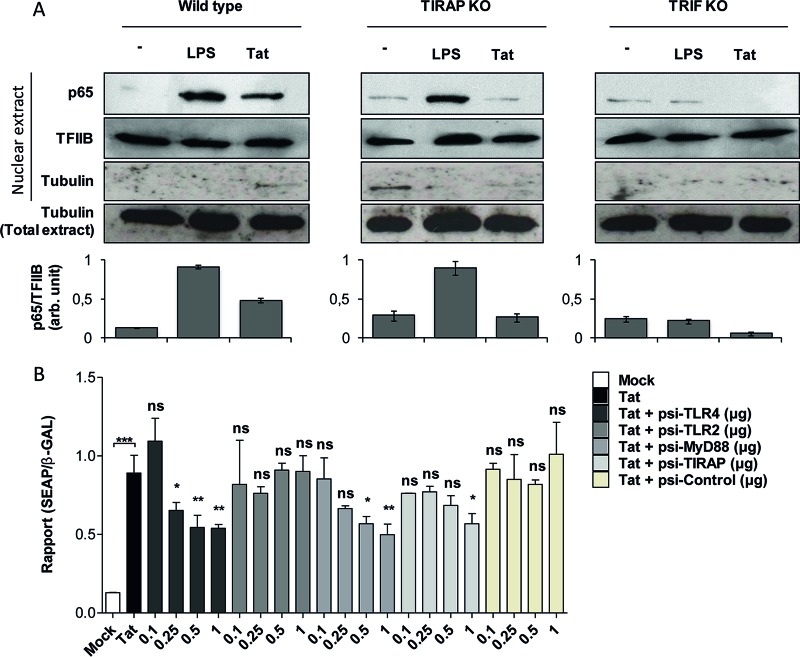
FIG 5.
HIV-1 Tat protein activates NF-κB downstream of TLR4-, MyD88-, TIRAP/MAL-, and TRIF-dependent pathways. (A) Primary mouse peritoneal macrophages isolated from WT, TIRAP/MAL−/−, or TRIF−/− mice were incubated with LPS (10 ng/ml) or the full-length Tat protein (100 nM) or left untreated (−) for 30 min. Cells were lysed, and the p65 subunit of NF-κB was detected in the nuclear fraction of the cells by Western blotting. TFIIB and tubulin were used as loading controls. Quantification of the bands obtained from 3 independent experiments was performed using ImageJ software. Data represent NF-κB (p65) expression in the nucleus normalized to TFIIB expression. (B) HEK cells expressing TLR4/MD2-CD14 were cotransfected with an NF-κB reporter plasmid (SEAP) together with the same amount of pORF-LacZ. Cells were also cotransfected with the indicated amounts (in micrograms) of plasmids encoding siRNAs targeting TLR4, TLR2, MyD88, and TIRAP/MAL or control siRNA or were left without transfection with siRNA (white bar, mock transfection; black bar, Tat stimulation). After 24 h of transfection, cells were left untreated (mock) or treated with Tat 1-45 (100 nM). Twenty-four hours after stimulation, NF-κB-driven SEAP reporter gene expression in the culture supernatants was measured. For normalization, cells were lysed and expression of β-galactosidase was quantified. The data represent means and SD for three independent experiments. Statistical significance was analyzed by one-way ANOVA followed by Bonferroni posttests and is marked as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant. All bars were compared to Tat-treated cells in the absence of cotransfection with any siRNA (black bars) unless otherwise specified.