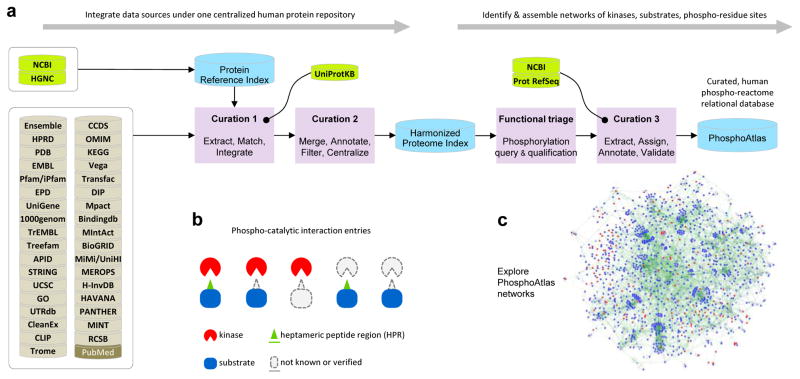
Figure 1. Phospho-reactome curation workflow: creation of a comprehensive and unified relational database that indexes human kinases and their cognate phosphorylation targets.
a. Resources, logical workflow, computational curation processes, and indexation steps are summarized. First, a Harmonized Proteome Index is created as a structured non-redundant repository of all known human proteins through curation steps 1 and 2, enabling the systematic extraction, harmonization and classification of public protein records. HGNC, NCBI and UniProtKB are used as critical databases that serve as blueprints to cross-reference and filter data and annotations extracted across 35 public databases. Second, PhosphoAtlas is build as a relational database of phosphorylation events. Curation ‘functional triage’ and step 3 identify which proteins are kinase enzymes or substrate proteins, how these proteins functionally interact with each other, and whether verifiable phosphorylatable residue sites and surrounding sequences can be defined. This establishes a comprehensive, curated dataset that maps human catalytic phospho-circuits, PhosphoAtlas.
b. Schematic representation of PhosphoAtlas entries. Five groups of complete or partial knowledge of phospho-catalytic interactions are shown. A majority of heptameric peptide regions (HPR’s) is centered on a phospho-residue site and stretch over 3 amino acids up and down, but for phospho-residues located at the N- or C-terminal of a protein, phospho-residues are displaced down or up the heptameric end portion of the protein.
c. PhosphoAtlas networks of kinase–substrate interactions can be explored (visual representation powered by Cytoscape (30)). Searchable CSV and XLS data files, and Cytoscape sessions, are downloadable upon registration at http://cancer.ucsf.edu/phosphoatlas.