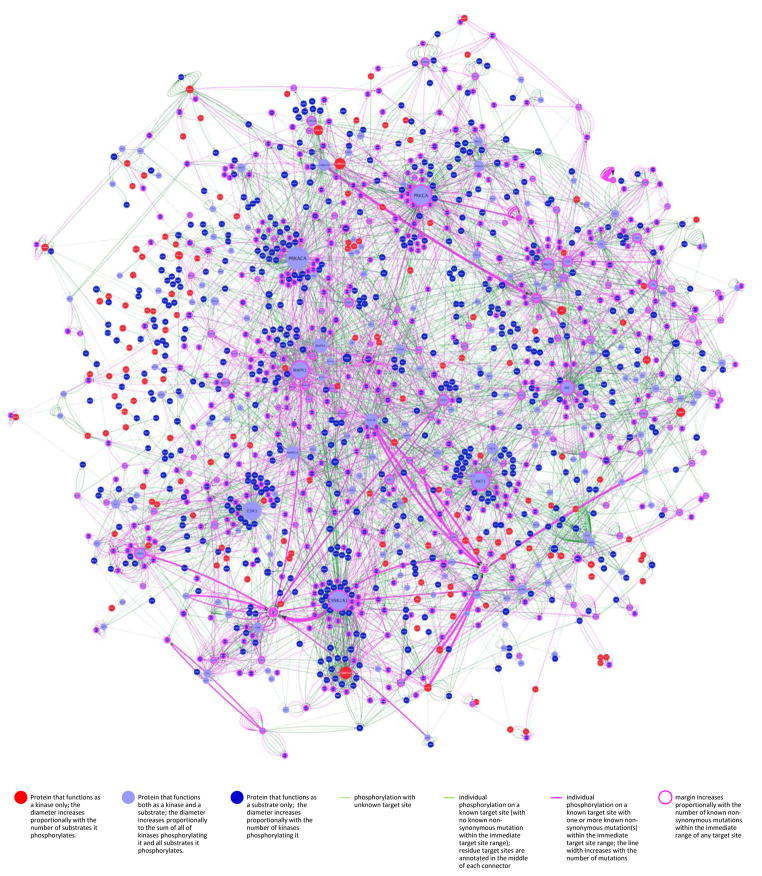
Figure 6. The tumor kinome: differential impact of cancer-associated mutations on kinase-substrate phospho-catalytic networks.
Data generated from cross-referencing PhosphoAtlas and COSMIC databases were used to render the differential impact –and potential convergence– of non-synonymous peptide target variants onto substrates of kinases. The complex heterogeneity of mutations across tumor genomes was resolved via the unbiased aggregation of mutations that only and selectively alter substrates’ HPRs. Purple connectors represent nsHPRv’s at the catalytic interface of kinases and substrates that are collapsed into more or less thick edge representing the prevalence of mutations occurring within one heptameric sequence. By overlaying both interconnectedness and mutational impact of tumors on the human phospho-reactome, the most variable nodes visually emerge from the otherwise rarely altered or not mutated network. The absence of any reported mutation for a number of prevalent nodes with high number of interactions is noticeable, such as CDK1, PRKACA, MAPK3, MAPK14, GSK3B, AKT3 or CSNK2A2. The phosphorylation networks were produced using the Cytoscape network analysis platform (30) and is available at http://cancer.ucsf.edu/phosphoatlas.