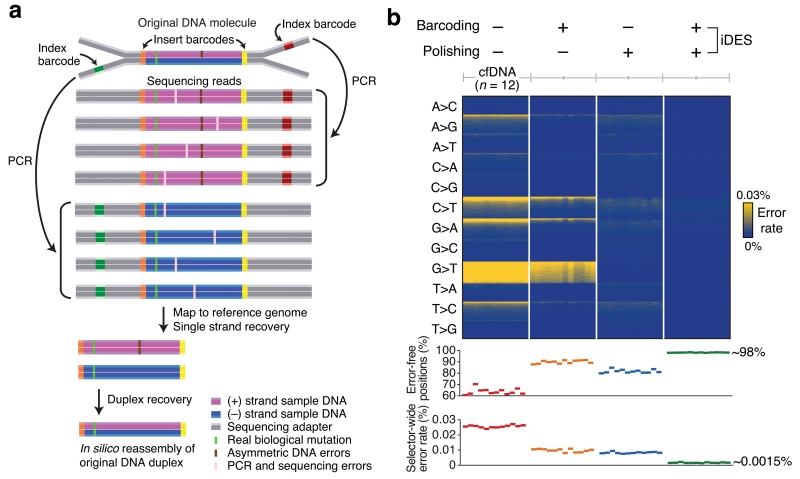
Figure 2. Development of integrated digital error suppression (iDES).
(a) Diagram depicting the use of CAPP-Seq barcode adapters to suppress errors. Here, CAPP-Seq adapters are ligated to a double-stranded (duplex) DNA molecule containing a real biological mutation in both strands as well as a non-replicated, asymmetric base change in only one strand (top). The combined application of insert and index barcodes allows for (i) error suppression and (ii) recovery of single stranded (center) and duplex (bottom) DNA molecules (Supplementary Fig. 1a, Methods). (b) Top: Heat map showing position-specific selector-wide error rates parceled into all possible base substitutions (rows) and organized by decreasing mean allele fractions (for each substitution type) across 12 cfDNA samples from healthy controls (columns; Supplementary Table 2). Background patterns are shown for different error suppression methods, including the combined application of barcoding and background polishing. Errors were defined as non-reference alleles excluding germline SNPs. Bottom: Selector-wide error metrics (Methods).