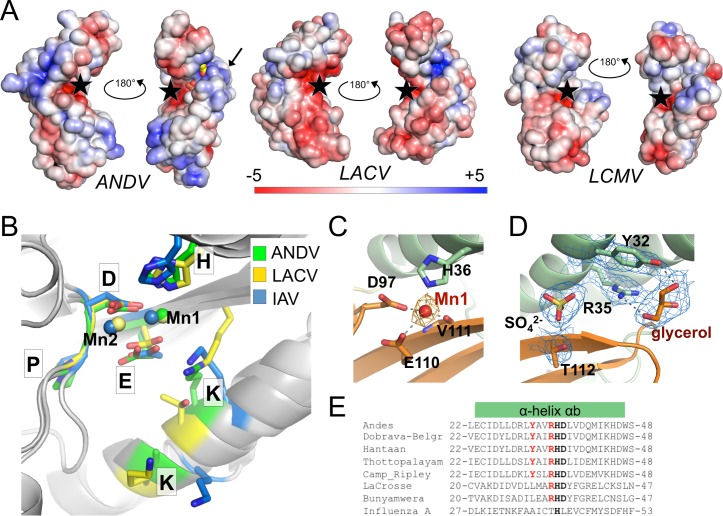
Fig 2. Surface charge distribution and active site arrangement of ANDV endonuclease compared to other cap-snatching endonucleases.
A, Electrostatic surface potential of the endonucleases of ANDV, LACV, and LCMV. The active site is marked with a black star, the location of helix αe in the ANDV structure is marked with a black arrow, sulfate and glycerol molecules bound in the basic groove of ANDV endonuclease are shown as spheres. The surface potential is shown from -5 KT/e in red to +5 KT/e in blue. B, Superimposition of the side chains of active site residues and the Mn2+ ions of ANDV, LACV, and IAV endonucleases. C, Manganese coordination in the active site of ANDV endonuclease. Metal coordinating residues are shown as sticks and coordination with dotted lines. The anomalous difference Fourier map for the manganese atom is shown as orange mesh at 3σ. D, Electron density for a sulfate and a glycerol molecule close to the active site of the ANDV endonuclease (2|Fo|-|Fc| map at 2σ is shown as blue mesh; ligands and coordinating side chains are shown as sticks). E, Tyr32 and Arg35 (in red), which coordinate the glycerol ligand in the ANDV L1–200 structure, are conserved in hantaviruses, but not present in IAV. LACV only possesses the Arginine. The catalytic His36 and the stabilizing Asp37 are shown in bold.