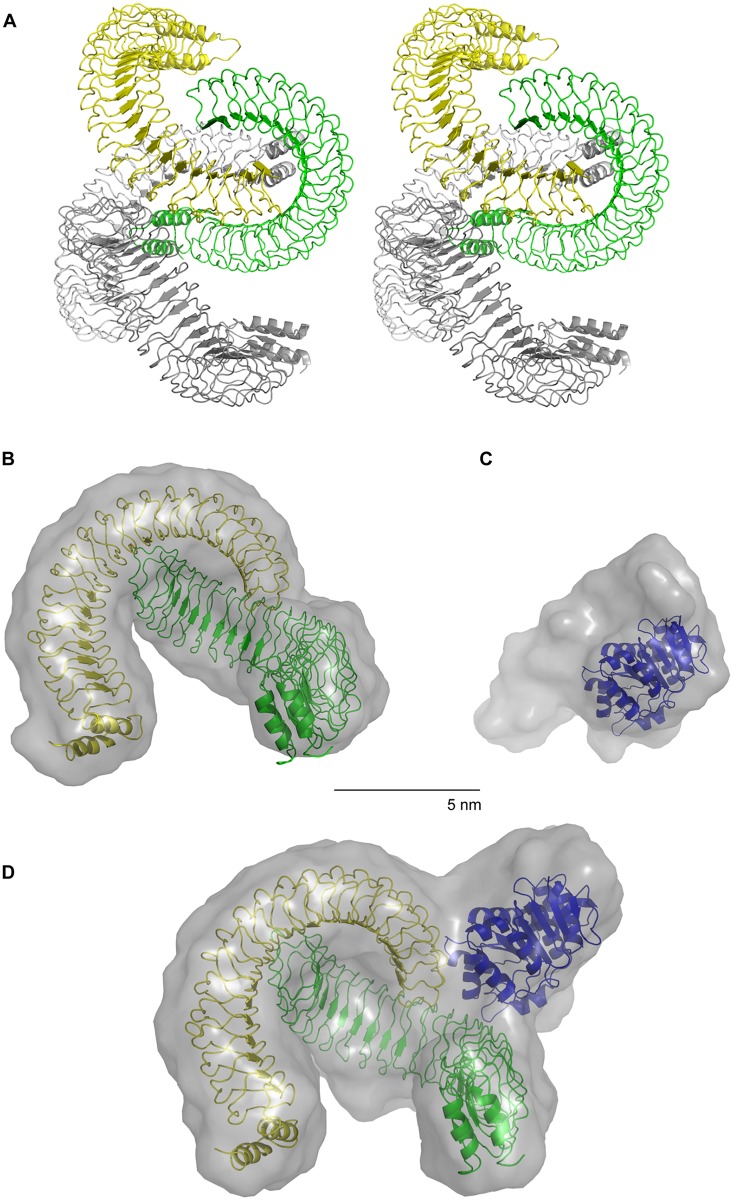
Fig 3. Crystal- and solution structure of YopM_34–481 and solution structures of DDX3_51–418 and the YopM_34-481/DDX3_51–418 complex.
A) YopM_34–481 was crystallized and its structure was solved (Methods; S2 Table). Stereo view shows ribbon representation of one asymmetric unit of the YopM_34–481 crystal (PDB code 4OW2). The asymmetric unit contains four molecules equivalent to two biological assemblies each represented by a dimer. YopM molecules of one dimer are colored in yellow and green and of the other dimer in light grey and grey. (B-D) The YopM_34–481 dimer, the DDX3_51–418 construct and the YopM_34-481/DDX3_51–418 complex were analyzed by small angle X-ray scattering (SAXS). B) The SASREF-model (Methods) of YopM_34–481 shown as transparent grey surface representation and the crystal structure of the YopM_34–481 dimer in ribbon representation are superimposed. C) The SASREF-model of DDX3_51–418 shown as transparent grey surface representation and the crystal structure of the ATPase domain of DDX3 (residues 167–418, PDB code 2I4I) are superimposed. D) The SASREF-model of the YopM_34-481/DDX3_51–418 complex shown as transparent grey surface representation is superimposed with the crystal structures of DDX3 (residues 167–418; [31] and the YopM_34–481 dimer (as presented in A). For experimental SAXS data see S2 Fig and S3 Table.