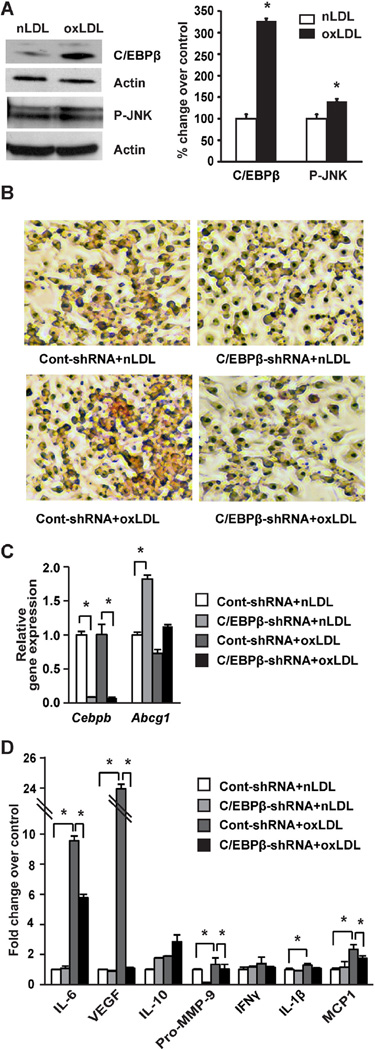
Fig. 5.
RAW264.7 macrophage cell treatment with oxLDL and effects of shRNA knockdown of C/EBPβ. A: RAW264.7 cells were treated with nLDL or oxLDL (20 µg/mL) for 24 h (n = 3 per experiment). Immunoblots and densitometric values for C/EBPβ (in nuclear fraction) and P-JNK (in cytosolic extract), with representative blots shown. Data represent the mean ± SEM, expressed as percent change over control after normalizing to actin. *P < 0.05 versus nLDL group as tested by Student t-test. B–D: RAW macrophage cells were transduced with control-shRNA (50 pfu/cell) or C/EBPβ shRNA (50 pfu/cell) for 24 h followed by treatment with nLDL or oxLDL (20 µg/mL) for an additional 24 h (n = 3 per experiment). B: Cells were fixed and stained with Oil Red O to detect lipid accumulation. C: Gene expression analysis by qPCR in RAW264.7 macrophage cells. Data are presented as mean ± SEM. *P < 0.05 as tested by two-way ANOVA followed by Tukey's post hoc test. D: Protein array data using conditioned medium collected from RAW cell experiments. Data are presented as mean ± SEM. *P < 0.05 as tested by two-way ANOVA followed by Tukey's post hoc test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)