Figure 1.
miRNA Design, Screening, and Viral-Vector Selection
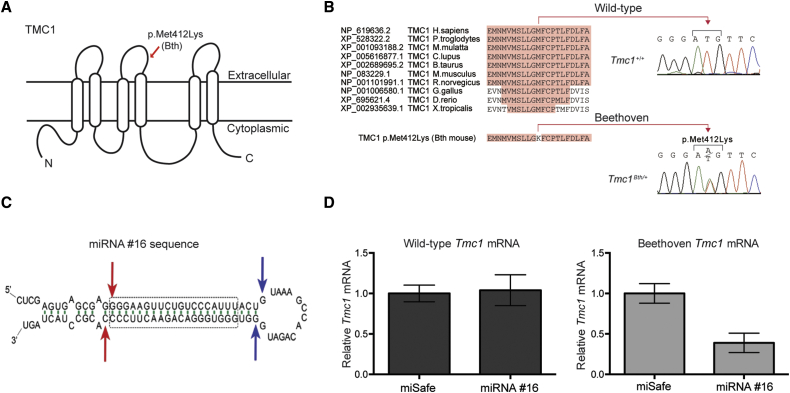
(A) Cartoon depiction of a possible configuration of TMC1 highlights the position of the Bth mutation.
(B) Multiple-sequence alignment shows conservation of Met412 in vertebrates and the Met412Lys change in the Tmc1Bth/+ mouse.
(C) siRNA sequence #16 embedded in an artificial miRNA scaffold. Of all miRNAs tested, #16 had the most specific and selective suppression of the mutant Tmc1 c.1235T>A allele. Blue and red arrows depict predicted Drosha and Dicer cleavage sites, respectively; the dashed box shows the core #16 sequence targeting the mutant Tmc1 variant.
(D) Real-time qPCR analysis of total RNA isolated from COS-7 cells cotransfected with constructs expressing both miRNA #16 and miSafe (a sequence specifically selected for its validated low off-targeting potential6) and either wild-type Tmc1 or mutant Tmc1 c.1235T>A. Relative mRNA expression levels were calculated with the ΔΔCt algorithm. Error bars represent the SD of three biological and nine technical replicates.