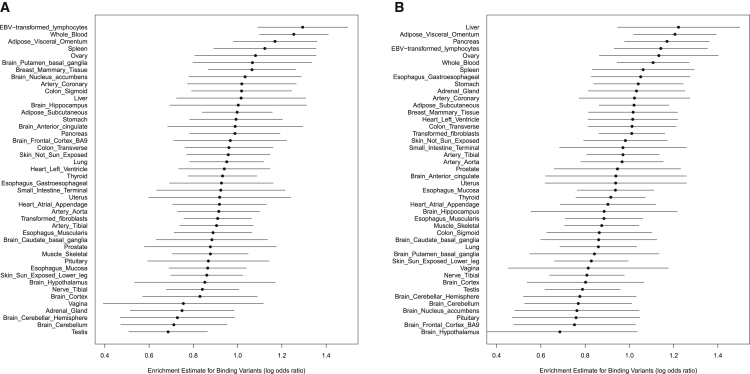
Figure 4.
Enrichment Estimates for Binding Variants in GTEx Tissues
The estimates in (A) are based on the annotations derived from the DNaseI data of the ENCODE LCLs, whereas the estimates in (B) are based on annotations derived from the ENCODE liver-related HepG2 DNaseI data. In each panel, we plot the point estimate of the enrichment parameter and its 95% confidence interval in each tissue. The tissues are ranked in descending order according to the magnitude of the point estimates. All estimates are obtained controlling for the SNP distance from TSS. All estimates are significantly far from 0 (at the 5% level). Interestingly, when the tissue and origin of the annotations match, the point estimates for enrichment are the highest.