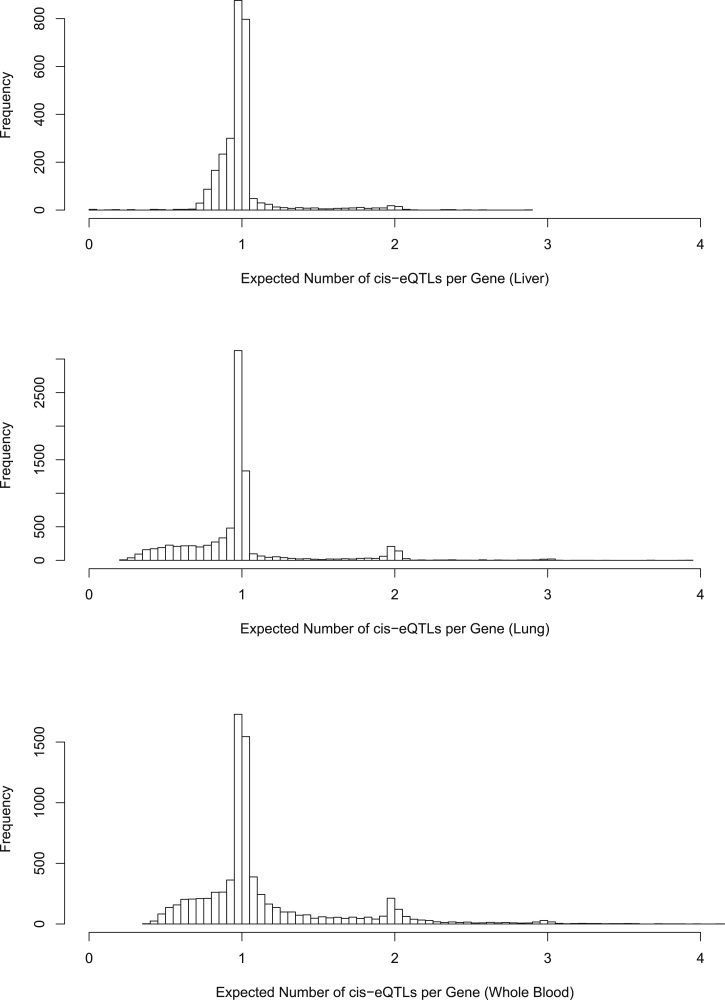
Figure 5.
Posterior Expected Number of cis-eQTL Signals per eGene in GTEx Liver, Lung, and Whole Blood Tissues
The top, middle, and bottom panels display the histogram of the posterior expected number of cis-eQTLs from all the eGenes in the liver, lung, and blood tissues, respectively. For most genes, we can identify only a single association signal. However, for a non-trivial number of eGenes, multiple independent association signals can be confidently identified by the adaptive DAP algorithm. The sample size is seemingly an important factor related to the ability to identify multiple independent signals in a cis region.