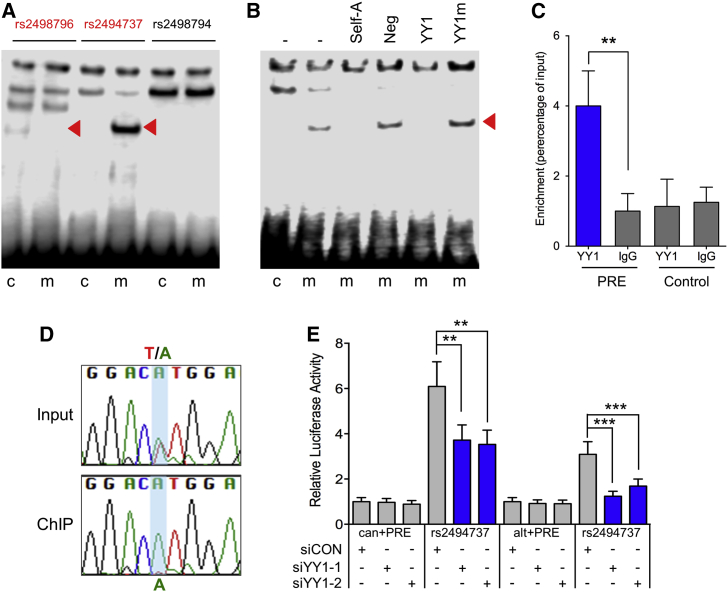
Figure 5.
The Risk Allele of rs2494737 Demonstrates Allele-Specific YY1 Binding
(A) EMSAs to detect allele-specific binding of nuclear proteins. Oligonucleotides were incubated with Ishikawa nuclear extracts. Red arrowheads show bands of different mobility detected between the common (C) and minor (risk-increasing) (m) alleles for the candidate causal SNPs.
(B) Oligonucleotides for SNP rs2494737 were incubated with Ishikawa nuclear extracts. Red arrowhead indicates the band that was competed for complex formation on the minor (m) allele. Competitor oligonucleotides are listed above each panel and were used at 100-fold molar excess: (−) no competitor; (Neg) a non-specific competitor; (YY1) consensus binding site; (YY1m) an identical oligonucleotide but with a mutated binding site.
(C) ChIP–qPCR on SNP rs2494737 in heterozygous Ishikawa cell lines. ChIP assays were performed with YY1 antibody or non-immune IgG, a region 3.2kb upstream of the predicted YY1-binding site served as a negative control (Control). Graphs represent two biological replicates. Error bars denote SD. P-values were determined with a two-tailed t-test (∗∗p < 0.01).
(D) Sanger sequencing of the PCR fragment generated using primers flanking SNP rs2494737 following YY1 ChIP-qPCR and the input DNA controls.
(E) Luciferase assays in Ishikawa cells shows the effect of YY1 siRNA silencing on the activity of the AKT1 canonical (can) and alternative (alt) promoter regions with the PRE containing the reference T allele (can+PRE; alt+PRE) or the risk A allele (rs2494737). Error bars denote 95% confidence intervals from three independent experiments performed in duplicate. P values were determined by two-way ANOVA followed by Dunnett’s multiple comparisons test (∗∗p < 0.01, ∗∗∗p < 0.001). The level of YY1 silencing is shown in Figure S8.