Fig. 5.
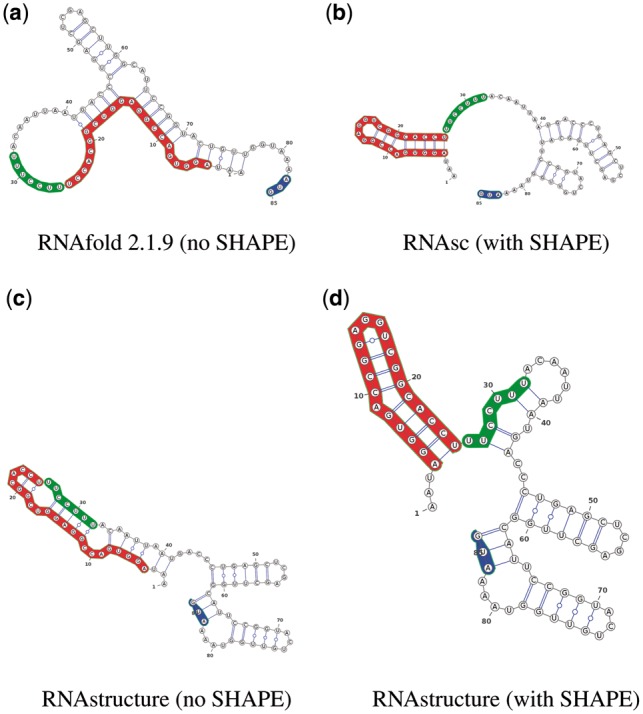
Secondary structure predictions of domain 5 of wild-type FMDV IRES, including the first functional AUG start codon, both with and without integration of probing data using selective -hydroxyl acylation analyzed by primer extension (SHAPE). (a) RNAfold 2.1.9, which does not incorporate SHAPE data. (b) RNAsc (Zarringhalam et al., 2012), which penalizes deviations from SHAPE data for both base paired and loop regions. (c) RNAstructure (Reuter and Mathews, 2010) without SHAPE data. (d) RNAstructure (Deigan et al., 2009), which penalizes deviations from SHAPE data only for base paired regions. The polypyrimidine tract (PPT) is unbound in only structures (a) from RNAfold 2.1.9 and (b) from RNAsc, compatible with the full IRES structure appearing in Figure 1 of Fernandez et al. (2011)
