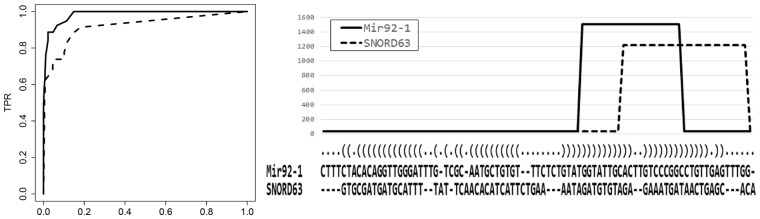
Fig. 2.
(Left.) ROC curves representing the true positive rates versus false positive rates based on clustering trees generated by SHARAKU and deepBlockAlign. The solid line represents the ROC curve of SHARAKU, and the dashed line represents the ROC curve of deepBlockAlign. (Right.) An alignment produced by SHARAKU for a pair of RNAs (Mir92-1 and SNORD63) in the simulated dataset with the annotations of RNA sequence alignment and the predicted secondary structure. The solid line represents the read coverages of Mir92-1, and the dashed line represents the read coverages of SNORD63