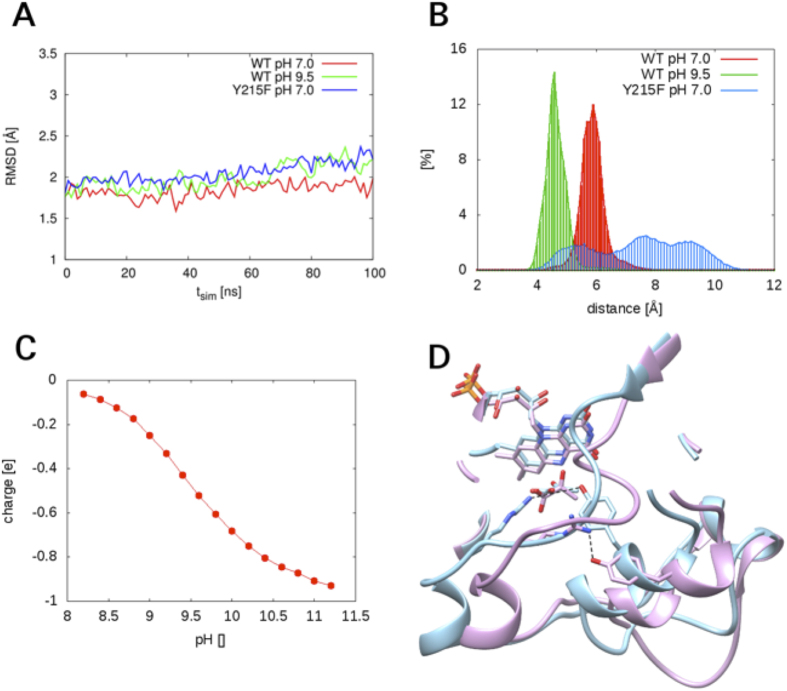
Figure 7. Results of MD simulations of wild-type and Y215F lactate oxidases are shown.
(A) R.m.s.d. traces from the 100-ns simulations performed using the enzyme crystal structure (PDB code 2E77) as reference. The experimental crystal structure was determined at pH 8.0. Note: a pre-equilibration time of 5 ns was used that is not shown in the graph. The r.m.s.d. traces therefore do not start at a value of zero. (B) The relative frequency distributions of distances between the guanidino Cζ of Arg181 and the phenyl Cζ of Tyr215/Phe215. (C) The calculated curve of pH-dependent ionization of Tyr215 in the (oxidized) enzyme-pyruvate complex of wild-type avLOX. (D) A superimpositioning of MD simulated structures of wild-type lactate oxidase at pH 7.0 (blue) and pH 9.5 (magenta). The structures shown are the final snapshots after 100 ns of MD simulation. The hydrogen bond between Arg181 and Tyr215 (set to be ionized at pH 9.5) is shown with a dashed line. The distance at pH 7.0 was 4.6 Å, that at pH 9.5 was only 2.9 Å.