Summary
Bacterial conjugation is the main mechanism responsible for the dissemination of antibiotic resistance genes. Hence, the search for specific conjugation inhibitors is paramount in the fight against the spread of these genes. In this pursuit, unsaturated fatty acids have been found to specifically inhibit bacterial conjugation. Despite the growing interest on these compounds, their mode of action and their specific target remain unknown. Here, we identified TrwD, a Type IV secretion traffic ATPase, as the molecular target for fatty acid-mediated inhibition of conjugation. Moreover, 2-alkynoic fatty acids, which are also potent inhibitors of bacterial conjugation, are also powerful inhibitors of the ATPase activity of TrwD. Characterization of the kinetic parameters of ATPase inhibition has led us to identify the catalytic mechanism by which fatty acids exert their activity. These results open a new avenue for the rational design of inhibitors of bacterial conjugation in the fight against the dissemination of antibiotic resistance genes.
Keywords: bacterial conjugation, traffic ATPases, bacterial secretion, inhibition
Graphical abstract
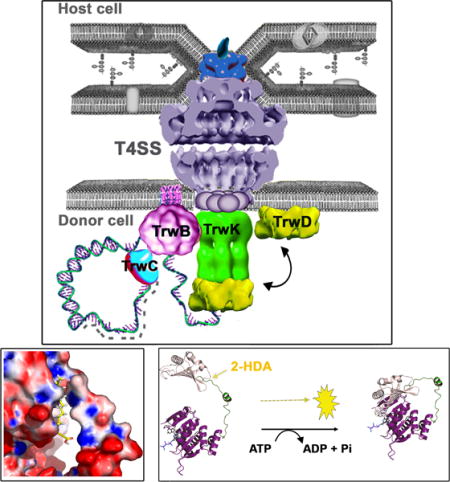
Introduction
Antibiotic resistance has become one of the most challenging problems in health care (WHO-report, 2014), as morbidity and mortality rates upon infection by multi-resistant pathogens have sharply increased over the last decades (Giske et al., 2009, Hawkey and Jones, 2009, Rice, 2009, Boucher et al., 2009). One of the main mechanisms whereby bacteria become resistant to antibiotics is the acquisition of antibiotic resistance genes by bacterial conjugation (Mazel and Davies, 1999, de la Cruz and Davies, 2000, Waters, 1999). Therefore, the search for specific conjugation inhibitors (COINs) is crucial for the control of the dissemination of antibiotic resistance genes. Most compounds reported to inhibit conjugation turned out to be unspecific growth inhibitors (Lujan et al., 2007, Hooper et al., 1989, Michel-Briand and Laporte, 1985, Leite et al., 2005, Conter et al., 2002). Bacterial conjugation has also been reported to be inhibited by the M13 phage coat protein g3p (Lin et al., 2011). However, the most promising results have been obtained with unsaturated fatty acids (uFAs), which specifically inhibit plasmid conjugation without inhibiting Escherichia coli growth (Fernández-Lopez et al., 2005, Getino et al., 2015).
Bacterial conjugation is powered by a set of ATPases that energize every single step in the conjugative process: plasmid relaxation and DNA unwinding, DNA transfer, pilus biogenesis and protein transport (for a recent review see (Cabezón et al., 2015)). These processes are catalyzedby TrwC, TrwB, TrwK and TrwD, which are the VirD2, VirD4, VirB4 and VirB11 homologues in the conjugative plasmid R388, respectively. TrwC is a protein that cleaves the DNA at the origin of transfer and presents relaxase and DNA helicase activities (reviewed in (Garcillán-Barcia et al., 2009, de la Cruz et al., 2010)). TrwB is a DNA-dependent ATPase that mediates the transfer of DNA to the secretion system (Tato et al., 2005). TrwK is an ATPase (Arechaga et al., 2008) that mediates the dislocation of the pilin molecules from the inner membrane during pilus biogenesis (Kerr and Christie, 2010). The fourth ATPase, TrwD, participates in pilus biogenesis (Kerr and Christie, 2010) and in DNA translocation (Atmakuri et al., 2004), acting as a molecular switch between pilus biogenesis and substrate transport (Ripoll-Rozada et al., 2013).
Here, we have analyzed the effect of the uFAs on the ATPase activity of all conjugative ATPases. We found that only TrwD, the VirB11 homologue, is inhibited by the same uFAs that inhibit bacterial conjugation in vivo. These uFAs did not affect significantly any other ATPase of the T4SS or the DNA mobilization machinery. TrwD (VirB11) belongs to a large family of hexameric AAA+ traffic ATPases, which includes proteins involved in Type II secretion and in Type IV pilus and flagellar biogenesis (Planet et al., 2001). All members of this superfamily are characterized by the presence of an N-terminal domain (NTD), required for membrane association, and a catalytic domain (CTD) connected by a linker region of variable length (Planet et al., 2001, Peña and Arechaga, 2013, Hare et al., 2006). Pivoting of the NTD relative to the CTD over the flexible linker has been proposed to be an essential step in ATP catalysis (Savvides et al., 2003, Yamagata and Tainer, 2007, Ripoll-Rozada et al., 2012). In this work, we have characterized the mechanism of inhibition of TrwD by uFAs. These compounds act as non-competitive inhibitors, with no effect on the affinity of the protein for ATP or ADP substrates. Blind docking of uFAs on the structural model of TrwD led to the identification of a region, comprising the NTD and the linker region of the protein, as the putative uFAs binding site. This result suggests a mechanism of inhibition that prevents the movement of the NTD over the CTD. As a consequence, VirB11 traffic ATPases become promising targets for the development of specific COINs based on uFA derivatives. This novel antimicrobial approach opens up new perspectives in the fight against the spread of antibiotic resistance genes.
Results
TrwD is the molecular target of linoleic acid inhibition
Unsaturated fatty acids (uFAs) are inhibitors of bacterial conjugation (Fernández-Lopez et al., 2005). It has been suggested that uFAs might target the conjugation machinery by affecting any of the ATPases associated with the type IV secretion system (T4SS) of donor cells. In order to determine the specific target of uFAs in our model system, the conjugative plasmid R388, the activity of the four ATPases involved in the conjugative process (TrwB, TrwC, TrwD and TrwK) was analyzed in vitro in the presence of 50 μM linoleic acid. At this concentration, 95 % reduction of the ATPase activity of TrwD was observed (Fig. 1), whereas no significant effect on the activity of any of the other three ATPases was detected. These results suggest that TrwD is a specific target of uFAs.
Figure 1. Effect of linoleic acid on the ATPase activity of conjugative ATPases.
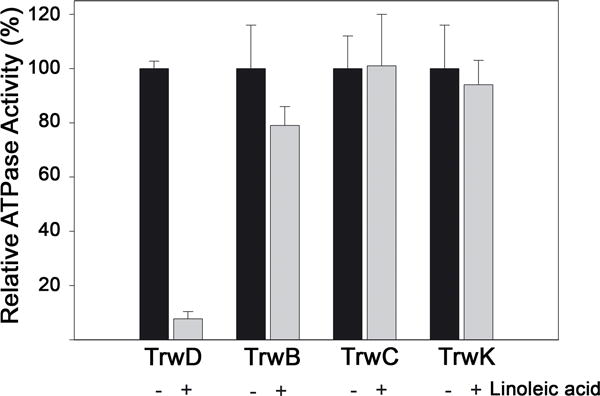
ATPase activity by each of the four ATPases that power conjugation (TrwC, TrwB, TrwK and TrwD) (2 μM) was measured in the absence or presence of linoleic acid (50 μM). As ATPase rates are different for each ATPase, hydrolysis activity is expressed as relative ATPase activity (error bars: SD).
Unsaturation of fatty acids is essential for TrwD ATPase inhibition
In contrast to uFAs, saturated fatty acids are not able to inhibit bacterial conjugation (Fernández-Lopez et al., 2005). In order to determine whether there was a correlation between the in vivo experiments and the in vitro analysis, we tested the effect of different types of fatty acids in TrwD ATPase activity. Oleic (C18:1(9)) and linoleic (C18:2 (9,12)) acids (cis-unsaturated C18 fatty acids, with one and two double bonds, respectively), previously identified as effective inhibitors of bacterial conjugation in vivo assays (Fernández-Lopez et al., 2005), were selected as uFAs. Lauric acid (C12:0) and Palmitic acid (C16:0), which are unable to inhibit significantly R388-mediated conjugation (Fernández-Lopez et al., 2005), were chosen as representative examples of saturated fatty acids. Stearic acid (C18:0) is not soluble in our experimental conditions and, therefore, it was not possible to test its effect. TrwD ATPase activity was measured in the presence of each fatty acid (50 μM). At this concentration, oleic and linoleic acids were able to inhibit more than 90% of TrwD ATPase activity whereas in the presence of saturated fatty acids no inhibition was observed (Fig. 2). These results strongly support that the presence of a double bond at C-9 is important for the TrwD ATPase inhibitory activity.
Figure 2. Effect of saturated and unsaturated fatty acids on TrwD ATPase activity.
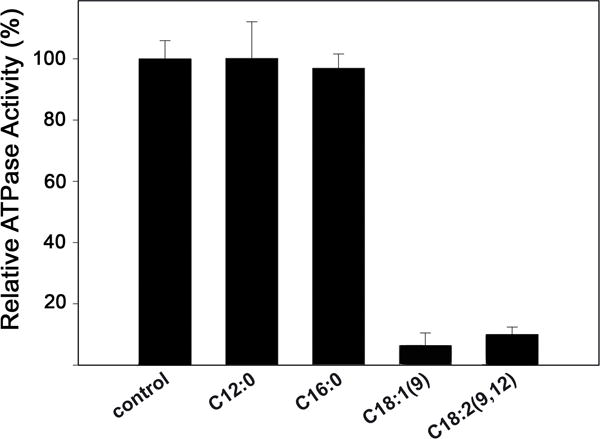
The ATPase activity of TrwD (2 μM) was measured in the presence of 50 μM lauric (C12:0), palmitic (C16:0), oleic (C18:1(9)), and linoleic (C18:2(9,12)) (error bars: SD).
2-alkynoic fatty acids are synthetic inhibitors of TrwD ATPase activity
Synthetic fatty acids, such as the 2-alkynoic fatty acids (2-aFAs), a class of acetylenic fatty acids with one triple bond between the C-2 and C-3 carbons of the alkyl chain, have been found to be effective in inhibiting bacterial conjugation (Getino et al., 2015). In particular, the three compounds found to be more effective in inhibiting bacterial conjugation were 2-hexadecynoic acid (2-HDA), 2-octadecynoic acid (2-ODA), and 2,6 hexadecadiynoic acid (2,6-HDA). Interestingly, alcohol (OH) or tetrahydropyranyl-ether (THP) derivatives of these compounds were unable to inhibit bacterial conjugation, suggesting an essential role of the carboxylic group in the mechanism of inhibition (Fig. 3A). Therefore, we decided to test whether TrwD was the molecular target of 2-aFAs, as in the case of the linoleic acid. Analysis of TrwD ATPase activity in the presence of 2-aFAs (50 μM) corroborated the results observed in vivo, as the same compounds that inhibited bacterial conjugation were also able to inhibit TrwD ATPase activity, whereas those with no effect in vivo, such as alcohol or tetrahydropyranyl-ethers derivatives, did not have any effect on the in vitro TrwD activity (Supplemental Fig S3). Moreover, the lack of inhibitory effect of these derivatives was still present even at concentrations as high as 500 μM (Fig. 3B). These results strongly reinforce the hypothesis that TrwD is the specific target of both, alkenoic and alkynoic fatty acids.
Figure 3. Effect of 2-alkynoic fatty acids and derivatives on TrwD ATPase activity and bacterial conjugation.
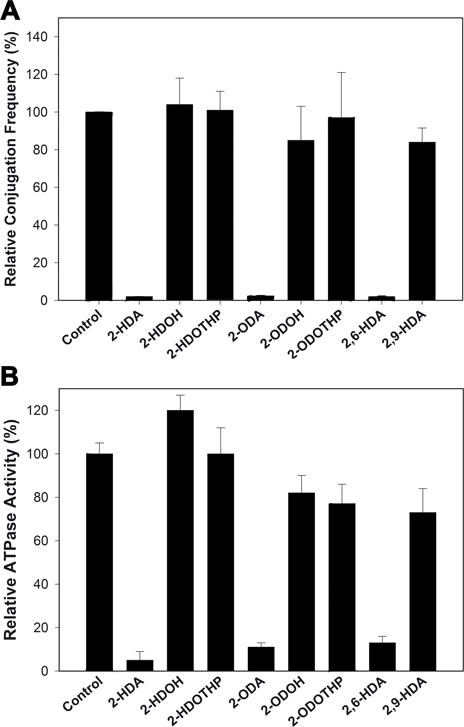
A; Bacterial conjugation was monitored in the presence of 2-alkynoic fatty acids derivatives (500 μM). B; ATPase activity by TrwD (2 μM) was tested in the presence of 500 μM of 2-octa (2-ODA) and 2-hexa-decynoic acids (2-HDA, 2,6-HDA and 2,9-HDA), and alcohol (-OH) or tetrahydropyranyl-ether (THP) derivatives. (error bars: SD).
Inhibition of ATP hydrolysis by fatty acids is non competitive
With the aim of better understanding the mechanism of inhibition of TrwD ATP hydrolysis by fatty acids, a characterization of the inhibition kinetics was carried out. First, ATPase activity rates of TrwD (2 μM) at increasing concentrations of linoleic acid, 2-HDA 2,6-HDA, or 2-ODA were determined (Fig. 4). Analysis of the kinetics of inhibition of TrwD ATPase activity by the four compounds showed a similar inhibition pattern. In all cases, data did not fit to a Michaelis-Menten inhibition kinetic curve, but to a sigmoidal Hill equation for inhibition, which suggested a cooperative effect in the inhibition kinetics. The apparent inhibition constants (Ki[app]) of linoleic acid, 2-HDA, 2-ODA and 2,6-HDA were 20.9 ± 1.6 μM, 29.7 ± 2.1 μM, 29.8 ± 4.1 μM and 43.8 ± 2.8 μM, respectively.
Figure 4. Determination of the kinetic parameters of inhibition by fatty acids.
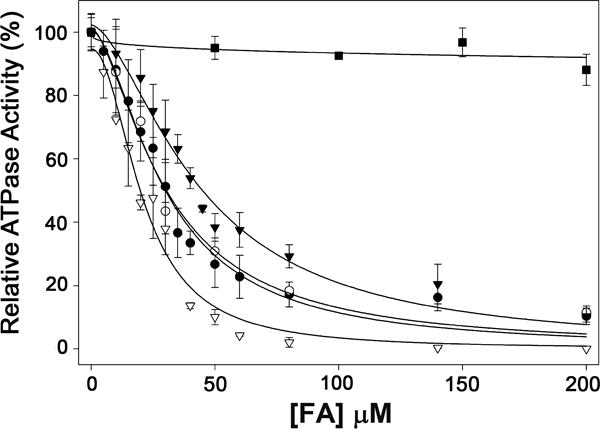
ATP hydrolysis by TrwD (2 μM) was measured at increasing concentrations of linoleic acid (white triangles), 2-HDA (black circles), 2-ODA (white circles), 2,6-HDA (black triangles) and palmitic acid (black squares). Data were fitted to a Hill inhibition equation (error bars: SD).
Further characterization of the mechanism of inhibition at steady-state was conducted in the presence of linoleic acid and 2-HDA. ATP turnover was measured at increasing concentrations of ATP in the presence or absence of fatty acids (Fig. 5A). The concentration of fatty acid was 21 μM linoleic acid or 30 μM 2-HDA, which correspond to their respective Ki[app]. In both cases the mechanism of inhibition was non-competitive, as Vmax was reduced without affecting the K0.5[ATP]. This result indicated that ATP binding was not affected by fatty acids.
Figure 5. Effect of fatty acids on nucleotide binding.
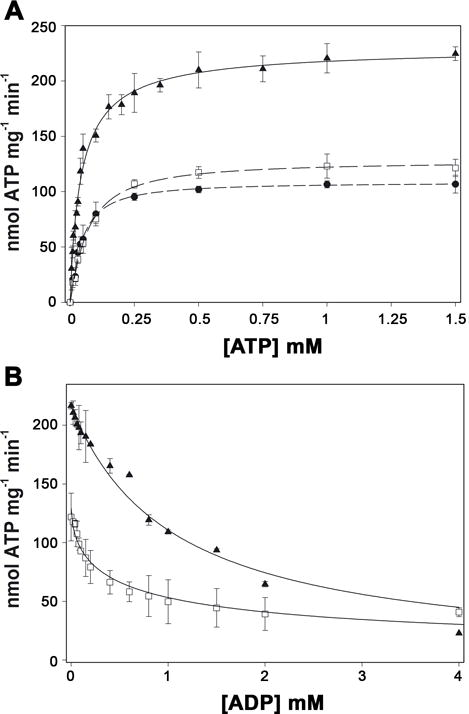
A, ATP hydrolysis by TrwD (2 μM) was measured in the presence of 21 μM of linoleic acid (black circles) and 30 μM 2-HDA (white squares), and in the absence of fatty acids (black triangles). Data were fitted to a Hill equation. The K0.5ATP was 66 μM in the presence of 2-HDA and 44 μM in the presence of linoleic acid and in the absence of added fatty acids. B, ATP hydrolysis by TrwD (2 μM) was measured at increasing concentrations of ADP in the presence of 30 μM 2-HDA (white squares) and in its absence (black triangles). Data were fitted as previously described (Ripoll-Rozada et al., 2012). The KdADP in the presence of 2-HDA and in the control were 51 μM and 45 μM, respectively. (error bars: SD).
In order to determine whether the inhibition by uFAs was caused by the stabilization of the enzyme in an ADP inhibited state, we analyzed the effect of ADP on TrwD ATPase activity in the presence and absence of 2-HDA (Fig. 5B). Interestingly, 2-HDA did not affect significantly the affinity of the enzyme for the ADP. In the presence of 2-HDA, the calculated KdADP was 51.7 ± 9.8 μM, and its absence this KdADP was 45.1 ± 4.1 μM. Altogether, these results show that the inhibitory effect of uFAs was not exerted by modifying the affinities of the enzyme for ATP or for the product, ADP.
Molecular docking of fatty acids into TrwD structure
Kinetic analysis of TrwD inhibition indicated that uFAs were binding to the protein in a site different from the nucleotide binding site, exerting the inhibition without affecting significantly ATP and ADP affinities. In order to explore putative binding sites for uFAs in TrwD, computer assisted analysis was performed. TrwD structural model was built by molecular threading using Brucella suis VirB11 (Hare et al., 2006) as a template, as previously described (Supplemental Fig. S4) (Ripoll-Rozada et al., 2012). Fatty acid ligands were retrieved from Pubchem repository (https://pubchem.ncbi.nlm.nih.gov/) and prepared for docking as described in Methods. Blind docking predictions using the EADock dihedral spacing sampling engine (Grosdidier et al., 2011) of the Swiss-dock server (http://www.swissdock.ch/) showed a region comprised by the NTD (residues 37–54) and the linker region (residues 118–125) of TrwD as the site with the highest probability for 2-HDA and linoleic acid binding (Fig. 6). All binding poses clustered at the same site when the structure of the hexameric form of TrwD was used as a target (Supplemental Fig. S5). The binding mode with the best energy and Full-Fitness is shown in Fig. 6. Similar results were obtained upon docking of 2-ODA and 2,6-HDA (Supplemental Fig. S7). Interestingly, tetrahydropyrane derivatives, such as 2-HDOTHP, although were able to fit in the same region, were adopting a different conformation due to the pyranyl ring (Supplemental Fig. S7).
Figure 6. Blind docking of fatty acids into the molecular model of TrwD.
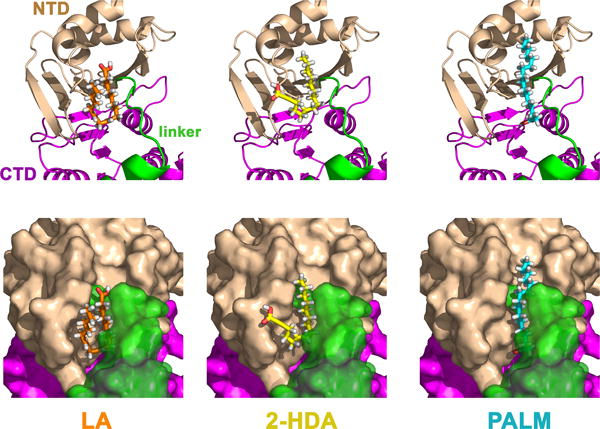
Blind docking predictions between a molecular model of TrwD (Ripoll-Rozada et al., 2012) and fatty acid ligands (linoleic acid (LA), 2-hexadecanoic acid (2-HDA), and palmitic acid (PALM)), were performed using the EADock dihedral spacing sampling engine of the Swiss-dock server (Grosdidier et al., 2011). Most of the binding poses clustered at a pocket localized at the interface between the N-terminal domain (NTD, wheat) and the linker region (green), which connects the NTD with the catalytic C-terminal domain (CTD, purple). The binding modes with the best energy and Full-Fitness are shown. Upper and bottom panels correspond to the same views in cartoon and surface representations, respectively. For clarity, transparency was applied to the linker region (green) in the surface representations (bottom panel).
Docking predictions with palmitic acid suggest that the binding site for this fatty acid is in the same region than the one found for linoleic acid and 2-HDA. However, uFAs and aFAs fill much better the binding pocket with higher Full-Fitness scores.
Partial proteolysis of TrwD in the presence of fatty acids
Blind docking search of 2-HDA and linoleic acid binding site identified the pocket comprised by the N-terminal domain (NTD) and the linker region of TrwD as the most likely site for binding (previous section). This linker region is essential during ATP catalysis, since it facilitates the movement of the NTD over the CTD in the catalytic mechanism of TrwD/VirB11 proteins. Therefore, ATPase inhibition by uFAs and aFAs might be caused by protein conformational changes upon fatty acid binding to this region. To test this hypothesis we analyzed the susceptibility of TrwD to partial digestion by papain in the presence of fatty acids. The unspecific protease papain is useful for delimiting structural domains in proteins, since it presents a low activity on stable secondary and tertiary structures (Karzai and McMacken, 1996). Thus, well-ordered structures would show low susceptibly to papain digestion. Incubation of TrwD with increasing concentrations of 2-HDA (1:10 to 1:100 TrwD:2-HDA molar ratios) resulted in high susceptibility to papain degradation (Fig. 7). In contrast, similar experiments performed in the presence of palmitic acid (1:100 TrwD : pamitic acid) did not affect the susceptibility of TrwD to papain digestion. These results suggest that 2-HDA binding induces an open conformation of TrwD, resulting in the exposition of protein domains to papain. Such a conformational change was not induced by palmitic acid, which is in accordance with the fact that saturated fatty acids do not exert any inhibitory effect.
Figure 7. Partial proteolysis of TrwD by papain.
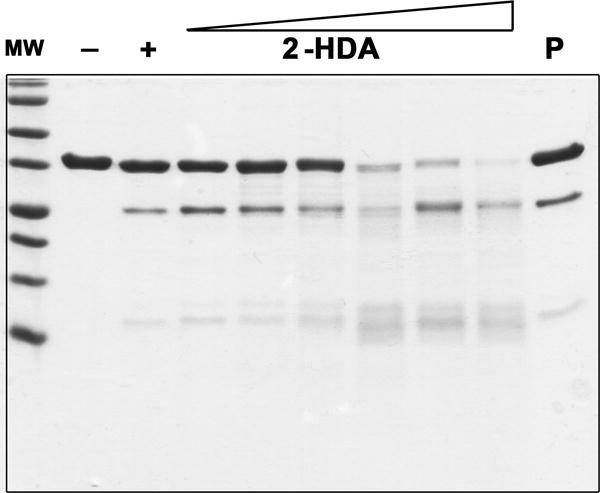
TrwD (lane -) was incubated with papain at a molar ratio of 1:80 (TrwD : papain) (lane +). Digestion performed in the presence of increasing concentrations of 2-HDA (1:10 to 1:100, TrwD : 2-HDA molar ratios) or palmitic acid (1:100, TrwD : palmitic acid molar ratio, lane P) revealed a different proteolysis pattern. Digestion was performed for 90 min at 25°C and proteolytic products were analysed by Tricine- SDS-PAGE (16.5% polyacrylamide gels).
Discussion
The search of conjugation inhibitors (COINs) is essential in the fight against multi-resistant bacteria. In this pursuit, high-throughput (HTS) methods have been developed to identify potential COINs. Although these methods have been widely used by pharmaceutical companies in hit identification (Mayr and Bojanic, 2009, Wu and Doberstein, 2006, Mishra et al., 2008), the finding of a molecular target for the structural-based design of new inhibitors could be a powerful tool in the development of new drugs, as it allows the investigation of the interactions between ligands and targets (Lounnas et al., 2013, Scapin, 2006). By using one of these HTS assays, uFAs have been identified as effective bacterial conjugation inhibitors (Fernández-Lopez et al., 2005). In that sense, the main goal of this work has been the identification and characterization of the molecular target for the inhibition by fatty acids.
uFAs are known to affect the function of some proteins that have a transient association with the bacterial membrane, such as the AAA+ ATPase DnaA (Yung and Kornberg, 1988). The ATPase activity of this DNA replication initiator is regulated by acidic phospholipids. The presence of saturated or unsaturated fatty acids on these phospholipids regulates the ADP-ATP exchange in the protein (Aranovich et al., 2006). Based on this, we decided to investigate the effect of uFAs on the ATPases involved in bacterial conjugation. In our model system, the conjugative plasmid R388, four ATPases provide the energy for the conjugative process: the relaxase TrwC, the coupling protein TrwB, TrwK and the traffic ATPase TrwD (Cabezón et al., 2015). Analysis of ATP hydrolysis rates by each of the four proteins in the presence of linoleic acid showed that only TrwD was inhibited under the tested conditions. This inhibitory effect was observed only in the presence of uFAs, such as oleic and linoleic acids, but not in the presence of saturated fatty acids like lauric or palmitic acids. These results correlate with previous observations, in which uFAs but not saturated ones were able to inhibit bacterial conjugation (Fernández-Lopez et al., 2005).
One of the down sides of uFAs is that they are prone to auto-oxidation, which could affect their effectiveness as deliverable COINs. In order to circumvent this, a series of synthetic uFAs containing triple bonds, which belong to the family of 2-alkynoic fatty acids (2-aFAs), were used as alternatives. These compounds are also good conjugation inhibitors (Getino et al., 2015). This family of fatty acids have been found to display antibacterial activity against Gram positive bacteria, such as Staphylococcus aereus and Bacillus cereus, and Gram negative bacteria like Klebsiella pneumoniae and Pseudomonas aeruginosa (Sanabria-Rios et al., 2014, Sanabria-Rios et al., 2015) but they not have any effect on E. coli growth (Konthikamee et al., 1982). In our experimental conditions these compounds did not have any effect on bacterial growth either. Thus, the observed inhibition was specific to bacterial conjugation, which is a desirable property, since they might help to control the dissemination of resistance genes without affecting the growth of commensal bacteria.
The carboxylic acid group of the fatty acid is essential to exert the inhibitory effect, since alcohol or tetrahydropyranyl-ether derivatives of these fatty acids were unable to inhibit TrwD ATPase activity (Fig. 3B). These results are in agreement with in vivo experiments in which these derivatives were also unable to inhibit bacterial conjugation (Fig. 3A). These experiments showed that 2-aFAs analogs, like 2-octadecynoic acid (2-ODA) and 2,6-hexadecynoic acid, were also efficient conjugation inhibitors. Fatty acid composition is also important, as unsaturation placed in C-9 instead of C-6, abolished the inhibitory effect of the hexadecynoic acid, both in vivo and in vitro experiments. Therefore, there is a direct correlation between the in vivo and in vitro data, as the same compounds able to inhibit bacterial conjugation were also capable of inhibiting TrwD ATPase activity, and vice versa, those unable to inhibit conjugation also failed to inhibit TrwD activity.
Kinetic analysis of TrwD ATPase inhibition by linoleic acid and 2-HDA showed similar Ki[app] parameters. In addition, ATP titration in the presence of either of the two compounds (at their respective MIC50) revealed no significant variation of the K0.5ATP when compared with the control, whereas the Vmax value was reduced by half, suggesting a non-competitive inhibition. The affinity for ADP did not change significantly, either. Altogether, these results suggest the presence of a binding site for uFAs and aFAs in TrwD different from the nucleotide binding site.
VirB11 proteins belong to a large superfamily of secretion ATPases (Planet et al., 2001), characterized by the presence of two distinct domains, NTD and CTD, connected by a flexible linker. This linker region is of particular importance during ATP catalysis, as movement of the NTD over the CTD has been suggested to be essential for the catalytic mechanism of TrwD/VirB11 (Savvides et al., 2003, Ripoll-Rozada et al., 2012). This catalytic mechanism has been proposed not only for VirB11 proteins but it has been extended to all members of the secretion ATPase superfamily (Yamagata and Tainer, 2007). According to this mechanism (Fig. 8), in the apo-state the enzyme could adopt two conformations, with the NTD alternating between an open and closed state, but only the open state is able to bind ATP. Upon ATP binding, the NTD closes over the CTD and ATP is hydrolyzed. The release of the γ-phosphate after ATP hydrolysis induces a conformational change to an open state that favors ADP release, so the cycle can resume. In a previous study (Ripoll-Rozada et al., 2012), we demonstrated that physiological concentrations of Mg2+ stabilizes the ADP-state of the enzyme (the affinity for ADP in the presence of Mg2+ was 100 times higher than in its absence), which, in turn, prevented the catalytic cycle to resume, thus reducing ATP turnover. In contrast, the results with fatty acids indicate a different mode of inhibition, as the affinty for ADP is hardly affected.
Figure 8. Mechanistic model of TrwD ATPase inhibition by fatty acids.
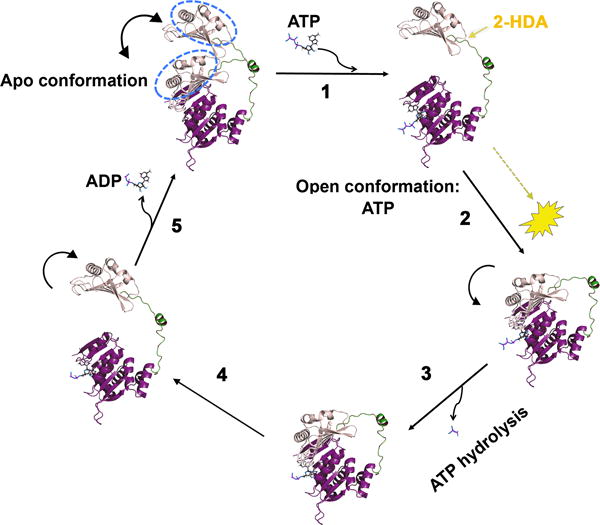
Proposed mechanism of the catalytic cycle of ATP hydrolysis (adapted from (Ripoll-Rozada et al., 2012)). According to this mechanism, which is shared by other members of the secretion ATPase superfamily (Yamagata and Tainer, 2007), in an apo- conformation, the protein can alternate between an open and a closed state, with the NTD pivoting over the CTD about the linker (step 1), but only the open conformation is able to bind ATP. Upon ATP binding, the NTD closes over the CTD (step 2) so ATP hydrolysis can take place (step 3). After ATP hydrolysis, the NTD opens up (step 4) allowing ADP release, so the cycle can resume (step 5). Considering that the binding affinities for ATP and ADP are not significantly affected by linoleic acid or 2-HDA, it is likely that the inhibitory effect of these COINs is exerted by preventing the movement of the NTD over the linker, which, in turn, results in inhibition of ATP hydrolysis.
As uFAs do not affect ATP or ADP significantly, they may act over another region of the protein. In fact, blind docking search of a putative binding site identified a pocket comprised by the N-terminal domain (NTD) and the linker region of TrwD with high probability to bind 2-HDA and linoleic acid (Fig. 6). It is likely, then, that binding of uFAs on this region results in the restriction of movement of the NTD over CTD, which, in turn, would result in a reduction of TrwD ATPase activity (Fig. 8). This binding pocket is not present in other VirB11 homologs, such as HP0525 from H. pylori or TraB from conjugative plasmid RP4 (Supplemental Fig. S4). The absence of this structural domain in TraB could explain previous reports in which no effect of these fatty acids on RP4-mediated conjugation was observed (Fernández-Lopez et al., 2005, Getino et al., 2015).
Further evidence of the conformational changes induced by uFAs was obtained by partial digestion of TrwD with papain, an unspecific protease widely used in the study of conformational modifications of proteins (Karzai and McMacken, 1996). In the presence of 2-HDA, the susceptibility of the protein to the digestion with papain increases (Fig. 7), which reinforces the idea of 2-HDA inducing conformational rearrangements in TrwD structure. Moreover, such conformational changes are not observed in the presence of palmitic acid, even at a 1:100 TrwD:palmitic acid molar ratio, which might explain the inability of saturated acids to inhibit TrwD activity. Although the blind docking search of the palmitic acid revealed a binding site similar to that found for uFAs and 2-HDA, the mode of binding is different. Unsaturated fatty acids acquire a cis- conformation which helps them to fill the cavity in the binding site, whereas saturated fatty acids have a rigid straight conformation. The finding of the binding site for uFAs at the NTD is also in accordance with previous data, which suggest that VirB11 proteins interact with the cytoplasmic site of the membrane through the NTD (Yeo et al., 2000). In addition, TrwD ATPase activity is stimulated by phospholipids (Rivas et al., 1997), and the protein has been shown to interact with phospholipids in lipid vesicles (Machón et al., 2002). Thus, it is very likely that uFAs and aFAs are occupying a binding site in TrwD, which is otherwise occupied by bacterial membrane phospholipids involved in the association of the protein to the membrane. In fact, E. coli cells treated with 2-HDA incorporate this fatty acid into the phospholipids (Sanabria-Rios, Getino and de la Cruz, unpublished observations). This result would make possible the in vivo interaction of 2-HDA with the membrane associated form of TrwD. Paradoxically, uFAs were also reported to inhibit conjugation of F-like plasmids (Getino et al., 2015) although no VirB11 homolog has yet been identified in F plasmids, begging the question of which is the target of COIN inhibition in F-like plasmids. Given the fact that F-like plasmids are narrow host range (i.e., appear only in Enterobacteriaceae), it is tempting to speculate that the VirB11 function is carried out by a traffic ATPase provided by the host.
In summary, in this work we have identified the conjugative T4SS traffic ATPase TrwD as the target for the inhibition of bacterial conjugation by fatty acids. The finding of conjugative traffic ATPases as potential targets for inhibiting bacterial conjugation opens a new avenue for the development of rational design drugs based on the interactions between this protein and potential inhibitors, which may help in the fight against the emergence of antibiotic multi-resistant bacteria. Future research should address the development of compounds with increased affinities for the target capable to inhibit conjugation irreversibility.
Experimental Procedures
Cloning, Overexpression and Purification of proteins
Cloning, overexpression and purification of TrwD, TrwC, TrwB and TrwK were carried out as described previously (Ripoll-Rozada et al., 2012, Tato et al., 2005, Arechaga et al., 2008, Grandoso et al., 1994). TrwC purification was modified as follows. After a P11 phosphocellulose column (Whatman), TrwC-enriched fractions were pooled and applied to a HiTrap SP-Sepharose (5ml) column (Amersham, GE), followed by a Mono S HR 5/5 (1 ml) column (Amersham, GE). Protein was eluted from both columns in a linear gradient of NaCl. Finally, the protein was concentrated and loaded onto a 200 HR Superdex column (Amersham, GE). Fractions were pooled and stored as described previously (Grandoso et al., 1994).
Chemicals and inhibitory compounds
Commercial fatty acids were purchased from Sigma-Aldrich (St. Louis, MO, USA). The 2-alkynoic fatty acids (2- aFAs) and derivatives were synthesized as described in (Carballeira et al., 2006, Carballeira et al., 2012). Alcohol, methyl ester and tetrahydropyranil ether compounds derivatives were obtained as described in (Sanabria-Rios et al., 2014).
ATP hydrolysis assays
Steady-state ATP hydrolysis activity was measured with the EnzCheck™ Kit (Invitrogen) in a UV-1800 spectrophotometer (Shimadzu). Inorganic phosphate (Pi) released after ATP hydrolysis was monitored as an increase of absorption at 360 nm for 10 min following manufacturer’s instructions and components: 0.2 mM 2-amino-6-mercapto-7-methylpurine riboside (MESG) and 1 unit/ml of purine nucleoside phosphorylase (PNP). A specific buffer was used for each ATPase to register ATP hydrolysis at optimal conditions. TrwD and TrwC ATP hydrolysis rates were determined in buffer DC (50 mM Tris-HCl pH 8.5, 75 mM potassium acetate, 10μM magnesium acetate and 10% glycerol (w/v)). ATP hydrolysis by TrwB and TrwK was measured in buffers B and K, respectively. Buffer B; 50mM Pipes-NaOH pH 6.2, 35 mM sodium chloride, 5mM magnesium acetate and 5% glycerol (w/v). Buffer K; 50mM Pipes-NaOH pH 6.45, 75 mM potassium acetate, 10mM magnesium acetate, 5% glycerol (w/v), 1 mM sodium acetate, 1mM DTT and 0.1 mM EDTA. Before starting the reaction by adding the corresponding ATPase, ATP and fatty acids diluted in DMSO were added to the concentrations indicated in the text.
Conjugation assays
Conjugation frequencies were estimated using the high-throughput conjugation method previously described (Getino et al., 2015). Briefly, E. coli DH5α and BL21(DE3) strains were used as donor and recipient strains, respectively. Cells were mixed in 1:1 ratio and spotted onto LB-agar plates with different compounds. Mating plates were incubated at 37 °C for 6 h and bacteria were resuspended in M9 broth. OD600 and GFP emission from transconjugant cells were measured to quantify R388 transfer.
Papain proteolysis
Limited papain digestions were performed as described in (Ripoll-Rozada et al., 2012) with some modifications. Papain digestion was performed at 25 °C in 20 mM Tris (pH 8.5), 75 mM AcK, 10 % (w/v) glycerol. Papain stocks were dissolved in the same buffer and activated by addition of 50 mM β-mercaptoethanol (37 °C, 30 min) just before use. TrwD (18 μM) was incubated for 15 min at 25 °C in the presence fatty acids (2-HDA or palmitic acid). Proteolysis was initiated by addition of papain from the activated stocks at 1:80 papain:TrwD molar ratios. The reaction was stopped after 90 min at 25 °C by addition of 100 μM E-64 inhibitor (Sigma). Proteolysis products were analyzed by Tricine-SDS-PAGE (16.5% polyacrylamide gels) followed by staining with Coomassie Brilliant Blue.
Molecular modeling and ligand docking
An atomic model of TrwD was generated by molecular threading using as template the atomic coordinates of B. suis VirB11 (2gza.pdb) (Hare et al., 2006), as previously described (Ripoll-Rozada et al., 2012). The structural coordinates of 2-hexadecynoic acid (2-HDA) and linoleic acid were retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Molecules were prepared for docking using the DockPrep tool of UCSF Chimera software package (Pettersen et al., 2004). This involved the addition of hydrogens, the replacement of incomplete side chains with Dunbrack rotamer library (Dunbrack, 2002), the removal of solvent water molecules, and the inclusion of partial charges using AMBERff12SB force field. Files containing the atomic coordinates of the target protein and the fatty acids were submitted to the SwissDock server (http://www.swissdock.ch/), which uses an EADock dihedral spacing sampling (DSS) engine for docking drug-like ligands into macromolecules (Grosdidier et al., 2011). Docking runs were blind performed over the entire molecule, without defining any specific region of the protein in order to prevent bias. Results were examined with UCSF chimera and outer clusters were ranked according to Full-Fitness (FF) score of Swissdock. Binding poses with the best FF score and minimal energy were finally selected. Structural molecular representations were rendered with PyMOL (DeLano, 2008). Control runs were performed with the atomic coordinates of VirB11 from Brucella suis (2gza.pdb) (Hare et al., 2006) and HP0525 from Helicobacter pylori (1g6o.pdb) (Yeo et al., 2000) as target molecules. Electrostatic potential maps were calculated with the PDB2PQR application (Dolinsky et al., 2007), using PROPKA for pKa calculations, and the resulting APBS files (Baker et al., 2001) were rendered with Pymol.
Supplementary Material
Acknowledgments
This work was supported by the Spanish Ministerio de Economía y Competitividad (MINECO) grants BFU2011-22874 (to E.C and I A) and BFU2014-55534 (to FDLC) and EU VII Framework Program projects 282004/FP7-HEALTH-2011-2.3.1-2 and 612146/ICT-2013-10 (to FDLC). DSR thanks the support of the National Center for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health through Grant Number 5P20GM103475-13 and the Inter American University of Puerto Rico.
Footnotes
The authors declare that they do not have any conflict of interest.
References
- Aranovich A, Gdalevsky GY, Cohen-Luria R, Fishov I, Parola AH. Membrane-catalyzed nucleotide exchange on DnaA. Effect of surface molecular crowding. J Biol Chem. 2006;281:12526–12534. doi: 10.1074/jbc.M510266200. [DOI] [PubMed] [Google Scholar]
- Arechaga I, Peña A, Zunzunegui S, del Carmen Fernandez-Alonso M, Rivas G, de la Cruz F. ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J Bacteriol. 2008;190:5472–5479. doi: 10.1128/JB.00321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2015;39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- Carballeira NM, Cartagena M, Sanabria D, Tasdemir D, Prada CF, Reguera RM, Balana-Fouce R. 2-Alkynoic fatty acids inhibit topoisomerase IB from Leishmania donovani. Bioorg Med Chem Lett. 2012;22:6185–6189. doi: 10.1016/j.bmcl.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballeira NM, Sanabria D, Cruz C, Parang K, Wan B, Franzblau S. 2,6-hexadecadiynoic acid and 2,6-nonadecadiynoic acid: novel synthesized acetylenic fatty acids as potent antifungal agents. Lipids. 2006;41:507–511. doi: 10.1007/s11745-006-5124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conter A, Sturny R, Gutierrez C, Cam K. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J Bacteriol. 2002;184:2850–2853. doi: 10.1128/JB.184.10.2850-2853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecuar Graphics System. DeLano Scientific LLC; Palo Alto, CA: 2008. [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack RL., Jr Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Fernández-Lopez R, Machon C, Longshaw CM, Martin S, Molin S, Zechner EL, Espinosa M, Lanka E, de la Cruz F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology. 2005;151:3517–3526. doi: 10.1099/mic.0.28216-0. [DOI] [PubMed] [Google Scholar]
- Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- Getino M, Campos J, Fernández-López R, Fernández A, Caballeira NM, Sanabria-Ríos DJ, De la Cruz F. 2-alkynoic fatty acids as chemically stable conjugation inhibitors. mBio. 2015;6:e01032–01015. doi: 10.1128/mBio.01032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giske CG, Sundsfjord AS, Kahlmeter G, Woodford N, Nordmann P, Paterson DL, Canton R, Walsh TR. Redefining extended-spectrum beta-lactamases: balancing science and clinical need. J Antimicrob Chemother. 2009;63:1–4. doi: 10.1093/jac/dkn444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandoso G, Llosa M, Zabala JC, de la Cruz F. Purification and biochemical characterization of TrwC, the helicase involved in plasmid R388 conjugal DNA transfer. Eur J Biochem. 1994;226:403–412. doi: 10.1111/j.1432-1033.1994.tb20065.x. [DOI] [PubMed] [Google Scholar]
- Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Bayliss R, Baron C, Waksman G. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol. 2006;360:56–66. doi: 10.1016/j.jmb.2006.04.060. [DOI] [PubMed] [Google Scholar]
- Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009;64:i3–10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- Hooper DC, Wolfson JS, Tung C, Souza KS, Swartz MN. Effects of inhibition of the B subunit of DNA gyrase on conjugation in Escherichia coli. J Bacteriol. 1989;171:2235–2237. doi: 10.1128/jb.171.4.2235-2237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- Kerr JE, Christie PJ. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol. 2010;192:4923–4934. doi: 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konthikamee W, Gilbertson JR, Langkamp H, Gershon H. Effect of 2-alkynoic acids on in vitro growth of bacterial and mammalian cells. Antimicrob Agents Chemother. 1982;22:805–809. doi: 10.1128/aac.22.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite JR, Silva LP, Rodrigues MI, Prates MV, Brand GD, Lacava BM, Azevedo RB, Bocca AL, Albuquerque S, Bloch C., Jr Phylloseptins: a novel class of anti-bacterial and anti-protozoan peptides from the Phyllomedusa genus. Peptides. 2005;26:565–573. doi: 10.1016/j.peptides.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Lin A, Jimenez J, Derr J, Vera P, Manapat ML, Esvelt KM, Villanueva L, Liu DR, Chen IA. Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLoS One. 2011;6:e19991. doi: 10.1371/journal.pone.0019991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounnas V, Ritschel T, Kelder J, McGuire R, Bywater RP, Foloppe N. Current progress in Structure-Based Rational Drug Design marks a new mindset in drug discovery. Comput Struct Biotechnol J. 2013;5:e201302011. doi: 10.5936/csbj.201302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc Natl Acad Sci U S A. 2007;104:12282–12287. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machón C, Rivas S, Albert A, Goñi FM, de la Cruz F. TrwD, the hexameric traffic ATPase encoded by plasmid R388, induces membrane destabilization and hemifusion of lipid vesicles. J Bacteriol. 2002;184:1661–1668. doi: 10.1128/JB.184.6.1661-1668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Mazel D, Davies J. Antibiotic resistance in microbes. Cell Mol Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel-Briand Y, Laporte JM. Inhibition of conjugal transfer of R plasmids by nitrofurans. J Gen Microbiol. 1985;131:2281–2284. doi: 10.1099/00221287-131-9-2281. [DOI] [PubMed] [Google Scholar]
- Mishra KP, Ganju L, Sairam M, Banerjee PK, Sawhney RC. A review of high throughput technology for the screening of natural products. Biomed Pharmacother. 2008;62:94–98. doi: 10.1016/j.biopha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Peña A, Arechaga I. Molecular motors in bacterial secretion. J Mol Microbiol Biotechnol. 2013;23:357–369. doi: 10.1159/000351360. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci U S A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB. The clinical consequences of antimicrobial resistance. Curr Opin Microbiol. 2009;12:476–481. doi: 10.1016/j.mib.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Ripoll-Rozada J, Peña A, Rivas S, Moro F, de la Cruz F, Cabezón E, Arechaga I. Regulation of the type IV secretion ATPase TrwD by magnesium: implications for catalytic mechanism of the secretion ATPase superfamily. J Biol Chem. 2012;287:17408–17414. doi: 10.1074/jbc.M112.357905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezón E. Functional Interactions of VirB11 Traffic ATPases with VirB4 and VirD4 Molecular Motors in Type IV Secretion Systems. J Bacteriol. 2013;195:4195–4201. doi: 10.1128/JB.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas S, Bolland S, Cabezón E, Goñi FM, de la Cruz F. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J Biol Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- Sanabria-Rios DJ, Rivera-Torres Y, Maldonado-Dominguez G, Dominguez I, Rios C, Diaz D, Rodriguez JW, Altieri-Rivera JS, Rios-Olivares E, Cintron G, Montano N, Carballeira NM. Antibacterial activity of 2-alkynoic fatty acids against multidrug-resistant bacteria. Chem Phys Lipids. 2014;178:84–91. doi: 10.1016/j.chemphyslip.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria-Rios DJ, Rivera-Torres Y, Rosario J, Gutierrez R, Torres-Garcia Y, Montano N, Ortiz-Soto G, Rios-Olivares E, Rodriguez JW, Carballeira NM. Chemical conjugation of 2-hexadecynoic acid to C5-curcumin enhances its antibacterial activity against multi-drug resistant bacteria. Bioorg Med Chem Lett. 2015;25:5067–5071. doi: 10.1016/j.bmcl.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapin G. Structural biology and drug discovery. Curr Pharm Des. 2006;12:2087–2097. doi: 10.2174/138161206777585201. [DOI] [PubMed] [Google Scholar]
- Tato I, Zunzunegui S, de la Cruz F, Cabezón E. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc Natl Acad Sci U S A. 2005;102:8156–8161. doi: 10.1073/pnas.0503402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters VL. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front Biosci. 1999;4:D433–456. doi: 10.2741/waters. [DOI] [PubMed] [Google Scholar]
- WHO-report. WHO: Antimicrobial Resistance. global report on surveillance 2014 [Google Scholar]
- Wu G, Doberstein SK. HTS technologies in biopharmaceutical discovery. Drug Discov Today. 2006;11:718–724. doi: 10.1016/j.drudis.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- Yung BY, Kornberg A. Membrane attachment activates dnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:7202–7205. doi: 10.1073/pnas.85.19.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


