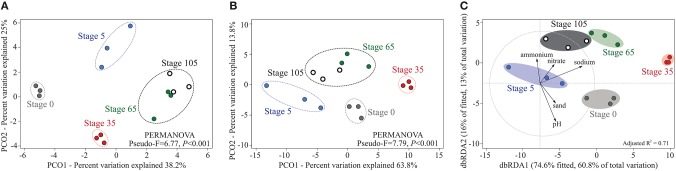
Figure 1.
Structure of the microbial communities along the successional gradient as determined by shotgun metagenomics. Plots illustrating distances between microbial communities in individual samples. (A) PCO based on complete functional community profiles (KO annotations). (B) PCO based on selected KOs involved in the N cycle (i.e., Nitrogen metabolism [PATH:ko00910]). Significant clusters are indicated by dashed lines (PERMANOVA, P < 0.05; see Supplementary Table 4 for details). (C) Distance-based redundancy analysis (dbRDA) illustrating the “best” fitting DistLM model (adjusted R2 = 0.71) containing forward selected predictor variables. Axis legends include % of variation explained by the fitted model and % of total variation explained by the axis.