Abstract
Recurrent outbreaks of infectious bursal disease (IBD) have become a burning problem to the poultry industry worldwide. Here, we performed genetic analysis of IBD virus (IBDV) field isolates from recent outbreaks in various poultry farms in India. The sequence analysis of IBDV VP2 hypervariable region revealed amino acid pattern similar to that of very virulent (222A, 242I, 253Q, 256I, 272I, 279D, 284A, 294I, 299S and 330S) and intermediate plus virulent (222A, 242I, 253Q, 256I, 272T, 279N, 284A, 294I, 299S and 330S) type whereas analysis of VP1 revealed presence of sequence similar to that of very virulent (61I, 145T) and unique (61I, 141I, 143D, 145S) type in field isolates. Among the eight field isolates, two isolates contained very virulent type VP2 and unique type VP1, three contained intermediate plus virulent type VP2 and unique type VP1 whereas five contained both VP2 and VP1 of very virulent type. The phylogenetic analysis based on VP2 nucleotide sequence showed clustering of all eight isolates close to known very virulent strains whereas based on VP1, five isolates formed unique cluster and three isolates were placed close to very virulent strains. The isolates forming unique VP1 cluster showed highest similarity with classical virulent IBDVs suggesting their possible evolution from segment B of non-very virulent IBDVs. Interestingly, these five isolates were responsible for outbreaks in four different farms located at three different geographic locations in India. These observations indicates genetic reassortment between segment A and segment B from co-infecting IBDV strains leading to emergence of very virulent strains and their widespread prevalence in Indian poultry farms. The presence of 272I and 279D in VP2 protein of five field isolates may explain possible cause of Gumboro intermediate plus vaccine failure in prevention of the outbreaks. However, mortality caused by other three strains which are antigenically similar to VP1 of intermediate plus vaccine strains could not be explained and the possible role of their unique VP1 in enhancing the pathogenesis needs to be investigated further.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-016-0306-z) contains supplementary material, which is available to authorized users.
Keywords: IBDV, Outbreaks, Poultry, India, Reassortment, Genetic drift
Introduction
Infectious bursal disease (IBD) is an acute and highly contagious viral disease affecting young chickens [7, 18]. The virus has a special predilection for the bursa of Fabricius, an important lymphoid organ in the young chicken, resulting in the immunosuppression and high mortality. The IBD virus (IBDV) belongs to the genus Avibirnavirus and is a member of the family Birnaviridae [9]. It is a non-enveloped virus consists of bi-segmented (segment A and B) double stranded RNA genome packaged in icosahedral capsid of 60 nm diameter [10]. Genome segment A is made of 3.2 kb whereas segment B is made of 2.8 kb linear double stranded RNA. Segment-A encodes four proteins encoded by two overlapping open reading frames. The larger ORF encodes a polyprotein which gets processed into three proteins VP2, VP3 and VP4 by the auto proteolytic action of viral protease VP4 [16] whereas smaller ORF encodes nonstructural VP5 protein [22]. VP2 protein forms outer capsid whereas VP3 protein forms the inner capsid. The segment-B encodes for VP1 protein having RNA dependent RNA polymerase (RdRp) activity [31]. VP2 is a major structural protein of the virus which elicits protective neutralizing antibodies and contains determinants responsible for causing antigenic variations. Although genome segment-A encoded proteins especially VP2 has been investigated in great detail as a major determinant of virulence, segment-B encoded VP1 protein has also been recently implicated for enhanced pathogenesis of virus through enhancing virus replication [11, 20]. Recently, genomic reassortants with segment-B have been found to cause emergence of pathogenic strains in China [32, 33].
IBDV serotype I strains are pathogenic posing great concern to the poultry industry whereas serotype II strains are avirulent for chickens. Based on the virulence properties, IBDVs are classified as attenuated (atIBDV), classical virulent (cvIBDV), antigenic variants (avIBDV) and very virulent (vvIBDV) subtypes [29]. Based on the conserved substitutions in the hypervariable region of VP2, the amino acids 222A, 256I, 279D, 284A, 294I and 299S were reported to be present in the all vvIBDV strains [5, 14, 25] and 222P, 256V, 279N, 284T, 294L and 299N were found to be conserved in atIBDV strains whereas the cvIBDV strains were reported to harbor the amino acids 249Q and 254G in VP2 hypervariable region [8, 30]. However, single amino acid substitutions such as I272T in ks strain were known to affect the virulence from very virulent to intermediate plus type (mb strain) [19]. The vvIBDV strains have spread to nearly every continent. In India, the vvIBDV was first reported in 1993 and despite of routine vaccination, the vvIBDV strains have been implicated in several field outbreaks. Genetic analysis of IBDV field isolates from India have described presence of amino acids variation in VP2 hypervariable region characteristic of very virulent type with unique aminoacid 212D > N substitution in some of the isolates [6, 17, 21, 27] and presence of unique amino acids substitutions in VP1gene [4]. These studies revealed circulation of very virulent IBDV subtypes in India and their continued evolution.
In the present study, we performed genetic analysis of segment-A encoded VP2 and segment-B encoded VP1, the two key determinants of viral pathogenesis, from recent field strains isolated from the various poultry farms in India. The birds in various poultry farms were vaccinated with intermediate vaccine at day 7 and intermediate plus vaccine at day 14 as well as between day 22 and 24 of age in layer birds whereas broiler birds received intermediate plus vaccine between 10 and 14 days of age. The onset of outbreaks were seen at the age of 3–4 weeks with mortality rate between 25–35 % in layer and 12–20 % in broiler birds. The genetic analysis of IBDV isolates from infected birds in our study demonstrates, genetic drift in VP2 hypervariable region resulting into virulent subtype and/or reassortment (genetic shift) with the genome segment B encoding unique VP1 as a possible cause for the emergence of recent outbreaks in the Indian poultry farms.
Materials and methods
Case history and sample collection
Bursa tissue samples were aseptically collected from chickens suspected to have died of infectious bursal disease from different poultry farms in India during the period from June 2015 to July 2015. The geographical locations of the farms are shown in Suppl. Figure 1. The infected birds were off fed, depressed with very high mortality, post mortem revealed the inflamed bursa filled with exudates and in many cases hemorrhagic bursa. The hemorrhagic lesions were observed in the muscles of thigh and breasts of the dead birds. The flocks of layer birds recovered eventually due to the post exposure immunity and became normal but the broiler birds suffered high morbidity due to post exposure immunosuppression complicated by E. coli infection and reduced weight gain. The tissue samples were collected and immediately transported to the laboratory in the presence of ice pack and frozen in −20 °C freezer until further processed for RNA isolation.
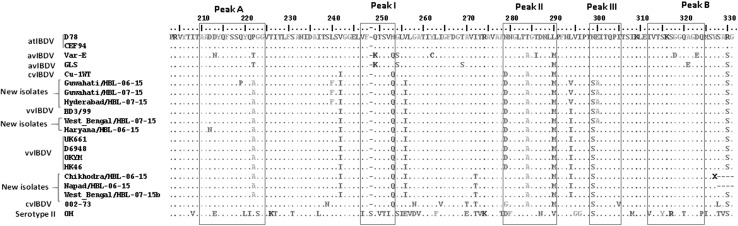
Fig. 1.
Amino acid sequence alignment of VP2 hypervariable region between amino acids 203–332 among the eight field isolates and 12 known IBDV strains. Major hydrophilic peaks A and B and minor hydrophilic peaks I, II and III are boxed
RNA isolation and RT-PCR
Approximately 100 mg of bursal tissue was homogenized in 1 ml of PBS using mortar pestle in the presence of glass beads. The homogenate was then clarified by centrifugation at 1300 rpm for 5 min. About 100 µl of the clarified homogenate was then subjected for RNA isolation using the Purelink RNA isolation kit (Invitrogen, Carlsbad, USA) as per the manufacturer’s instructions. The isolated RNA was subjected to RT-PCR using Qiagen one-step RT-PCR kit (Qiagen, Hilden, Germany) and gene specific primers. We targeted VP2 hypervariable region spanning 210–330 amino acids using VP2 forward TCACCGTCCTCAGCTTAC and VP2 reverse TCAGGATTTGGGATCAGC primers and VP1 N-terminus region spanning 10-260 amino acids using VP1 forward GCTCCTCTTCTTGATGATTC and VP1 reverse GTTYGAKATGGCTATCATCTC primers for identification of genetic variations and phylogenetic analysis. The PCR reaction was performed in 25 µl reaction mixture containing 5 µl of 5× buffer, 1 µl of enzyme mix, 1 µl of RNA and 10 pmol of VP1 or VP2 specific forward and reverse primers. The PCR reaction was initially subjected to 50 °C for 30 min for reverse transcription followed by 95 °C for 15 min for polymerase activation/template denaturation and 40 amplification cycles at 95 °C for 30 s, 52 °C for 30 s and 72 °C for 1 min with final extension of 72 °C for 5 min. The specific amplification of expected size PCR product was analyzed by agarose gel electrophoresis.
Sequencing and data analysis
The amplified PCR product was purified by purelink PCR purification columns (Invitrogen, USA) as per the manufacturer’s instructions. The PCR amplicons were sequenced at Xcelris labs, Ahmedabad by ABI3730xl capillary sequencer using BigDye terminator 3.1 cycle sequencing kit and gene specific forward and reverse primers as per the manufacturer’s instructions. Sequencing data were verified by manually inspecting the chromatogram using the sequencing analysis software v5.2 (Applied Biosystems, USA). The PCR amplification and sequencing of IBDV intermediate plus vaccine strains viz. Hester GI+, Ventri GIplus, Abic/MB and B2K and IBDV intermediate vaccine strains viz. Georgia intermediate and Baxendale intermediate were performed as described earlier. The nucleotide sequences of field isolates were aligned with the sequence of the known IBDV subtypes or IBDV vaccine strains using ClustalW software. The aligned sequences were downloaded and further analyzed using BioEdit software v7.2.3 [13]. The nucleotide sequences were translated to amino acids and further analyzed for the variations in the field isolates compared to sequences of known IBDV subtypes (Table 1) or sequences of vaccine strains (sequence data not shown). The phylogenetic analysis of field isolates with known IBDV subtypes or vaccine strains was performed with the help of MEGA6 software [28] using neighbor-joining method [26] and 1000 bootstrap replicates. The phylogenetic analysis of nucleotide sequences of field isolates with reported Indian field isolates were performed by MOLE-BLAST program of NCBI using the entrez query as India and default parameters. The phylogenetic tree was downloaded and further edited using the MEGA6 software.
Table 1.
Unique amino acids substitutions in VP1 and VP2 protein among IBDV virulent and attenuated strains
| Strain | Virulence Phenotype | VP2 | VP2 genotype | VP1 | VP1 genotype | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 222 | 242 | 249 | 253 | 254 | 256 | 272 | 279 | 284 | 294 | 299 | 330 | 61 | 141 | 143 | 145 | ||||
| Guwahati/HBL-06-15 | VV | A | I | Q | Q | G | I | I | D | A | V | S | S | VV | I | V | E | T | VV |
| Guwahati/HBL-07-15 | VV | A | I | Q | Q | G | I | I | D | A | V | S | S | VV | I | V | E | T | VV |
| Hyderabad/HBL-07-15 | VV | A | I | Q | Q | G | I | I | D | A | V | S | S | VV | I | V | E | T | VV |
| West Bengal/HBL-07-15 | VV | A | I | Q | Q | G | I | I | D | A | I | S | S | VV | I | I | D | S | Unique |
| Haryana/HBL-06-15 | VV | A | I | Q | Q | G | I | I | D | A | I | S | S | VV | I | I | D | S | Unique |
| Chikhodra/HBL-06-15 | VV | A | I | Q | Q | G | I | T | N | A | I | S | – | IPV | I | I | D | S | Unique |
| Napad/HBL-06-15 | VV | A | I | Q | Q | G | I | T | N | A | I | S | – | IPV | I | I | D | S | Unique |
| West Bengal/HBL-07-15b | VV | A | I | Q | Q | G | I | T | N | A | I | S | S | IPV | I | I | D | S | Unique |
| BD3/99 | VV | A | I | Q | Q | G | I | I | D | A | I | S | S | VV | I | V | E | T | VV |
| UK661 | VV | A | I | Q | Q | G | I | I | D | A | I | S | S | VV | I | V | E | T | VV |
| D6948 | VV | A | I | Q | Q | G | I | I | D | A | I | S | S | VV | I | V | E | T | VV |
| OKYM | VV | A | I | Q | Q | G | I | I | D | A | I | S | S | VV | I | V | E | T | VV |
| HK46 | VV | A | I | Q | Q | G | I | I | D | A | I | S | S | VV | I | V | E | T | VV |
| Cu-1WT | CV | P | I | Q | Q | G | V | I | D | A | I | N | S | CV | V | V | E | N | Non-VV |
| 002-73 | CV | P | V | Q | Q | G | V | T | G | A | L | S | S | CV | I | V | E | T | VV |
| D78 | AT | P | V | Q | H | G | V | I | N | T | L | N | R | AT | V | V | E | N | Non-VV |
| CEF94 | AT | P | V | Q | H | G | V | I | N | T | L | N | R | AT | V | V | E | N | Non-VV |
| GLS | AV | T | V | K | H | S | V | I | N | T | L | N | S | AV | V | V | E | N | Non-VV |
| Var-E | AV | T | V | K | Q | S | V | I | N | A | L | N | S | AV | V | V | E | N | Non-VV |
| OH (Serotype II) | NP | P | V | Q | H | S | E | T | D | T | L | S | S | NP | V | V | E | N | Non-VV |
Amino acids highlighted in italic are known to enhance virulence
VV very virulent, CV classical virulent, AT attenuated, AV antigenic variant, IPV intermediate plus virulence, NP non pathogenic
Nucleotide accession numbers
The VP1 and VP2 gene sequences generated in the study were deposited in GenBank under the accession numbers KT630841 to KT630856.
Results
VP2 hypervariable region and VP1 B-marker region of field isolates
Recurrent IBD outbreaks in the vaccinated flocks have been observed very frequently in several poultry farms from different geographic locations in India. Recently, we observed high mortality in the 3–4 week old chickens despite of routine vaccination with IBD intermediate and intermediate plus vaccines. In order to investigate possible genetic drift or reassortment leading to immune evasion in vaccinated birds, here we analyzed VP2 hypervariable region and VP1 B-marker region of IBD virus [2].
The bursa samples collected from eight birds belonging to different farms suspected to have died of IBDV infection during the period of June–July 2015 showed typical clinical signs of inflamed and hemorrhagic lesions. The PCR amplification of VP1 and VP2 gene segments using the RNA isolated from bursal tissues by one step RT-PCR confirmed the infection of IBDV. The RT-PCR amplified VP1 and VP2 gene segments were further sequenced and analyzed for nucleotide and amino acids variations. The nucleotide and deduced amino acid sequence suggested presence of VP2 signature sequence typical of very virulent strains in five of the eight field viruses isolated from different farms, however, there was significant variations among the different isolates suggesting prevalence of heterogeneous population of pathogenic isolates (Table 1). The detail clinical signs in the infected birds along with the identified VP1 and VP2 genotypes are presented in Suppl. Table 1.
Sequence comparison of field isolates with known IBDV subtypes and vaccine strains
Multiple sequence alignment of the VP2 hypervariable region from the IBDV isolates suggested their similarity with the known very virulent strains reported earlier (Fig. 1) whereas alignment of VP1 N-terminus region suggested presence of novel sequence variations in the five of the eight isolates (Fig. 2). The VP2 hypervariable region revealed presence of amino acids typical of the virulent strains i.e. 222A, 242I, 253Q, 256I, 272I, 279D, 284A, 294I, 299S and 330S in majority of the isolates (Guwahati/HBL-06-15, Guwahati/HBL-07-15, Hyderabad/HBL-07-15, West Bengal/HBL-07-15, Haryana/HBL-06-15) with few exceptions which includes presence of 272T and 279N in 3 isolates (Chikhodra/HBL-06-15, Napad/HBL-06-15, West Bengal/HBL-07-15b). We also observed novel substitutions L240F and L294V in 3 isolates (Guwahati/HBL-06-15, Guwahati/HBL-07-15, Hyderabad/HBL-07-15) and Q219P in Guwahati/HBL-06-15 isolate with very virulent genotype. The 3 isolates (Chikhodra/HBL-06-15, Napad/HBL-06-15, West Bengal/HBL-07-15b) which showed presence of 272T and 279 N showed VP2 genetic similarity with the intermediate plus vaccine strains. Based on the hydrophilicity and probability of surface exposure of amino acids in VP2 hypervariable region, hydrophilic regions were reported as major hydrophilic peak A (aa 210–225) and B (aa 312–324) and minor hydrophilic peaks I (aa 247–254), II (aa 281–292) and III (aa 299–305) [1, 12]. The field isolates were found to possess amino acids substitutions in the major peaks A and B and minor peaks I and II compared to the attenuated strains, however, substitutions were also observed in the regions excluding the hydrophilic peaks. The amino acids sequence alignment of VP2 hypervariable region of field isolates with vaccine strains (Suppl. Fig. 2a) showed higher similarity of all field isolates with the intermediate plus vaccine strains with few substitutions which includes 272I and 279D in 5 isolates, 245F and 294Vin three isolates and 300A in 4 isolates.
Fig. 2.
Amino acid sequence alignment of VP1 N-terminus region between amino acids 9–268 among the 8 field isolates and 12 known IBDV strains. Novel sequence variations identified in the field isolates are boxed
VP1 N-terminus sequencing revealed presence of 61I and 145T, similar to as reported for very virulent type in three of the eight isolates (Guwahati/HBL-06-15, Guwahati/HBL-07-15, Hyderabad/HBL-07-15). These three isolates also contained unique but neutral amino acid substitution K251R in VP1. Interestingly, other five field isolates (West Bengal/HBL-07-15, Haryana/HBL-06-15, Chikhodra/HBL-06-15, Napad/HBL-06-15, West Bengal/HBL-07-15b) showed unique amino acid substitutions such as 141I, 143D and 145S in VP1 N-terminus. Such amino acid variations have not been reported so far in any of the IBDV isolates. The amino acids alignment of VP1 of field isolates with vaccine strains (Suppl. Fig. 2b) revealed 96T and 161D in all field isolates similar to that of intermediate vaccine strains, 145T, 146D and 147N in three isolates viz. Guwahati/HBL-06-15, Guwahati/HBL-07-15, Hyderabad/HBL-07-15 similar to that of intermediate plus but differing from intermediate vaccine strains whereas 5 field isolates (West Bengal/HBL-07-15, Haryana/HBL-06-15, Chikhodra/HBL-06-15, Napad/HBL-06-15, West Bengal/HBL-07-15b) showed unique amino acid substitutions such as 141I, 143D and 145S not present in VP1 of both type of vaccine strains.
Phylogenetic analysis of field isolates with known IBDV subtypes and vaccine strains
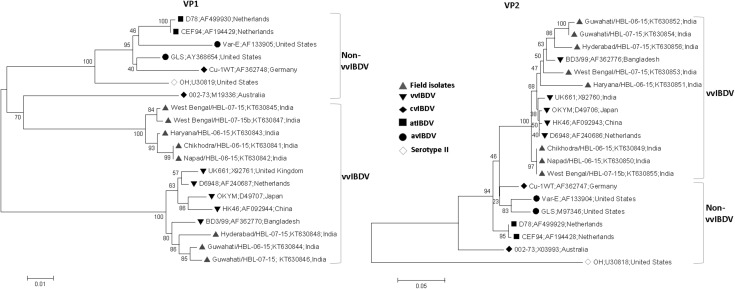
The phylogenetic analysis of VP2 hypervariable region showed clustering of the all eight isolates along with the known very virulent strains whereas VP1 phylogeny revealed distinct clustering of five of the eight isolates (West Bengal/HBL-07-15, Haryana/HBL-06-15, Chikhodra/HBL-06-15, Napad/HBL-06-15, West Bengal/HBL-07-15b) separate from virulent and non-virulent strains whereas 3 isolates (Guwahati/HBL-06-15, Guwahati/HBL-07-15, Hyderabad/HBL-07-15) showed clustering with the virulent strains (Fig. 3). Interestingly, these five isolates forming unique cluster based on VP1 showed close similarity of >98 % among themselves whereas of about 89 % similarity with 002–73 and GLS strains by pairwise distance analysis (Suppl. Table 2). The VP1 of remaining three isolates (Guwahati/HBL-06-15, Guwahati/HBL-07-15, Hyderabad/HBL-07-15) shared 97–98 % similarity with BD3/99, HK46, D6948, OKYM and UK661. The pairwise distance analysis of VP2 showed >99 % identity between Chikhodra/HBL-06-15, Napad/HBL-06-15 and West Bengal/HBL-07-15b isolates and >99 % identity between two Guwahati isolates. The Haryana/HBL-06-15, Hyderabad/HBL-07-15 and West Bengal/HBL-07-15 isolates showed >3 % divergence among each other as well as with other field strains. The Haryana/HBL-06-15, Hyderabad/HBL-07-15, West Bengal/HBL-07-15 and two Guwahati isolates shared 94–97 % nucleotide sequence identity with BD3/99, HK46, D6948, OKYM and UK661 strains (Suppl. Table 3).
Fig. 3.
Phylogenetic analysis of IBDV field isolates and known IBDV strains based on the VP1 B-marker region and VP2 hypervariable region. Tree was constructed by Neighbor-Joining method using sequences of 8 field strains, and 12 known IBDV strains of different subtypes. Values at the nodes indicates bootstraps probability as determined by 1000 re-sampling. vvIBDV: very virulent IBDV, cvIBDV: classical virulent IBDV, atIBDV: attenuated IBDV, avIBDV: antigenic variant IBDV. Serotype II is a non-pathogenic IBDV strains. Name of the IBDV isolates, their accession numbers and country of origin are given in the order
Phylogenetic analysis of field isolates with reported Indian isolates and vaccine strains
The phylogenetic analysis of VP2 sequence of the field isolates with reported Indian IBDV isolates suggested clustering with virulent field strains reported from the nearby geographical region (Suppl. Fig. 3a) whereas 5 of the 8 isolates showed unique type VP1 as evidenced by absence of closely related strains reported so far in India (Suppl. Fig. 3b). Phylogenetic analysis of field isolates with vaccine strains showed clustering of all isolates near intermediate plus vaccine strains based on VP2 sequence (Suppl. Fig. 4a) whereas field isolates carrying unique type VP1 placed close to intermediate vaccine strains whereas isolates with very virulent type VP1 were placed with intermediate plus vaccine strains (Suppl. Fig. 4b).
Discussion
Infectious bursal disease poses biggest threat to the poultry industry worldwide. Despite of routine vaccination, recurrent outbreaks have been observed in young chickens. Recent IBD outbreaks in various poultry farms at different geographic locations in India had resulted in high mortality which prompted us to investigate the underlying cause of vaccine failure. Based on recent observations, it is suggested that both genome segments of IBDV contributes to the pathogenesis. The genetic drift in the immunoprotective VP2 antigen and reassortment with the more virulent genome segment B leading to emergence of very virulent strains have been reported recently [1, 11, 32–34].
Considering the importance of both genome segment in determining virulence, we analyzed hypervariable region of VP2 responsible for antigenic variation as well as VP1 N-terminus region which is recently defined as B-marker region [2] for differentiation of virulent and non-virulent isolates. Analysis of VP2 hypervariable region suggested presence of virulence determining amino acids 222A, 242I, 253Q, 256I, 284A, and 299S [3, 23, 24, 32] in the all field isolates. The amino acids 272I, 279D and 294I which are also implicated in virulence have been observed in five of the eight isolates whereas three isolates were found to carry 272T and 279N which were observed in the attenuated vaccine strains. Three isolates were found to contain novel substitutions L240F and L294V which has not been reported for either virulent or attenuated strains. The functional significance of these amino acid substitutions in determining the virulence need to be ascertained. Among the eight field isolates, Chikhodra/HBL-06-15, Napad/HBL-06-15 and West Bengal/HBL-07-15b shared >99 % identity and two Guwahati isolates shared 99 % identity at nucleotide level suggesting their common origin whereas remaining three isolates differed by >4 % among themselves and shared identity of 94–97 % with BD3/99, HK46, D6948, OKYM and UK661 strains suggesting possible genetic drift in the VP2 hypervariable region of virulent parent strains. The D212N substitution which was found to be common in north China [35] and has been reported in India [6, 17] was also observed in one of the field isolates Haryana/HBL-06-15 in our study. The close clustering of VP2 sequence of field isolates with previously reported field strains of India suggests their evolution from circulating field strains. The presence of amino acid substitutions in the VP2 hypervariable region compared to intermediate and intermediate plus vaccine strains may partly contribute to failure of protection from vaccination in these farms.
The analysis of VP1 B-marker region suggested presence of 61I in all the isolates whereas 145T in three of the 8 isolates as reported for VP1 of very virulent types [34]. Interestingly, five of the eight isolates were found to contain novel amino acids substitutions 141I, 143D and 145S which are not reported for any of the IBDV strains so far. These amino acids substitutions were found to be common in five of the eight field strains isolated from 4 different farms. The phylogenetic analysis based on VP1 B-marker region suggested distinct clustering of these five isolates. At nucleotide level, these five isolates shared >98 % identity but differed significantly with the rest of the field isolates and known IBDV strains by >11–15 %. The phylogenetic analysis of VP1 with Indian field and vaccine strains revealed that none of the previous studies have reported VP1 of unique type identified in present study. These finding indicates that the five of the eight field isolates are reassortants with unique genome segment-B which is possibly appear to have descended from 002–73 related classical virulent strains which shared about 89 % sequence similarity, the highest among the known strains. VP1 of other three field isolates showed 97–98 % identity at nucleotide level with BD3/99, HK46, D6948, OKYM and UK661 suggesting their evolution from virulent parent strains. VP1 sequence analysis of IBDV field isolates from India reported high genetic and amino acids similarity with BD3/99, OKYM and UK661 strains [15]. Our finding provides support for the genetic reassortment of segment B with the segment A of co-infecting viral strains and emergence of more virulent strains as reported recently [32, 33]. Clinically, all field isolates led to severe pathogenicity whereas mortality pattern was higher in the layer birds compared to broilers. The possible implications of each VP1 and VP2 genotype on clinical phenotype thus need to be experimentally evaluated.
The present study on genetic analysis of VP1 and VP2 region of IBDV field isolates revealed widespread prevalence of genetic reassortants in Indian poultry farms. The majority of field isolates were found to possess VP2 gene similar to that of very virulent type with presence of unique amino acids substitutions in some of the isolates. The identification of unique amino acid pattern in the viral RNA polymerase VP1 in the present study and its significant genetic divergence from reported strains suggests evolution and adaptation of genome segment B and its reassortment with segment A of virulent or vaccine strains leading to emergence of new strains. These emerging isolates may overcome the immune barrier and lead to altered pathogenicity or replicative potential. Further study on their ability to cause disease in experimental birds and analysis of their virulence determinants would provide greater understanding of risks posed by these emerging isolates and will help to design future vaccination strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Authors wish to thank Hester Biosciences Ltd, India for providing necessary funding and infrastructure to support the present study.
Contributor Information
Amrutlal K. Patel, Phone: +91-79-2644-5106, Email: amrutkpatel@gmail.com
Joy K. Pal, Phone: +91-79-2644-5106, Email: jk.pal@hester.in
References
- 1.Adamu J, Owoade AA, Abdu PA, Kazeem HM, Fatihu MY. Characterization of field and vaccine infectious bursal disease viruses from Nigeria revealing possible virulence and regional markers in the VP2 minor hydrophilic peaks. Avian Pathol. 2013;42:420–433. doi: 10.1080/03079457.2013.822055. [DOI] [PubMed] [Google Scholar]
- 2.Alfonso-Morales A, Rios L, Martinez-Perez O, Dolz R, Valle R, Perera CL, Bertran K, Frias MT, Ganges L, de Arce HD, Majo N, Nunez JI, Perez LJ. Evaluation of a phylogenetic marker based on genomic segment B of infectious bursal disease virus: facilitating a feasible incorporation of this segment to the molecular epidemiology studies for this viral agent. PLoS One. 2015;10:e0125853. doi: 10.1371/journal.pone.0125853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bais MV, Kataria RS, Tiwari AK, Indervesh, Suryanarayana VV, Das SK. Sequence analysis of segment a of a field virus isolate from an outbreak of infectious bursal disease in India. Acta Virol. 2003;47:73–77. [PubMed] [Google Scholar]
- 4.Bais MV, Kataria RS, Tiwari AK, Viswas KN, Reddy AV, Prasad N. Sequence analysis of an Indian field isolate of infectious bursal disease virus shows six unique amino acid changes in the VP1 gene. Vet Res Commun. 2004;28:641–646. doi: 10.1023/B:VERC.0000042876.09951.b2. [DOI] [PubMed] [Google Scholar]
- 5.Banda A, Villegas P. Genetic characterization of very virulent infectious bursal disease viruses from Latin America. Avian Dis. 2004;48:540–549. doi: 10.1637/7157-12304R. [DOI] [PubMed] [Google Scholar]
- 6.Barathidasan R, Singh S, Kumar MA, Desingu P, Palanivelu M, Singh M, Dhama K. Recurrent outbreaks of infecous bursal disease (IBD) in a layer farm caused by very virulent IBD virus (vvIBDV) in India: pathology and molecular analysis. S Asian J Exp Biol. 2013;3:200–206. [Google Scholar]
- 7.Berg TPVD. Acute infectious bursal disease in poultry: a review. Avian Pathol. 2000;29:175–194. doi: 10.1080/03079450050045431. [DOI] [PubMed] [Google Scholar]
- 8.Brandt M, Yao K, Liu M, Heckert RA, Vakharia VN. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J Virol. 2001;75:11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas B, Kibenge F, Leong J, Mundt E, Vakharia V. Birnaviridae, Virus taxonomy: classification and nomenclature of viruses. In: Eighth report of the International Committee on Taxonomy of Viruses Elsevier/Academic Press, London, UK; 2005, pp. 561–569.
- 10.Dobos P, Hill BJ, Hallett R, Kells DT, Becht H, Teninges D. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J Virol. 1979;32:593–605. doi: 10.1128/jvi.32.2.593-605.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escaffre O, Le Nouen C, Amelot M, Ambroggio X, Ogden KM, Guionie O, Toquin D, Muller H, Islam MR, Eterradossi N. Both genome segments contribute to the pathogenicity of very virulent infectious bursal disease virus. J Virol. 2013;87:2767–2780. doi: 10.1128/JVI.02360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghazali MS, Ahmad-Raus R, Amin NM, Shohaimi SA, Bakar FDA, Zainuddin NB, Muhammad NAAN. Genomic analysis and comparison of very virulent infectious bursal disease virus (vvIBDV) affecting Malaysian poultry chickens with other IBDV strains. Ann Biol Res. 2013;4:92–104. [Google Scholar]
- 13.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 14.Hoque MM, Omar AR, Chong L, Hair-Bejo M, Aini I. Pathogenicity of Ssp I-positive infectious bursal disease virus and molecular characterization of the VP2 hypervariable region. Avian Pathol. 2001;30:369–380. doi: 10.1080/03079450120066377. [DOI] [PubMed] [Google Scholar]
- 15.Indervesh, Tiwari AK, Kataria RS, Viswas KN, Bais MV, Suryanarayana VV. Isolates of infectious bursal disease virus from India are of very virulent phenotype. Acta Virol. 2003;47:173–177. [PubMed] [Google Scholar]
- 16.Jagadish MN, Staton VJ, Hudson PJ, Azad AA. Birnavirus precursor polyprotein is processed in Escherichia coli by its own virus-encoded polypeptide. J Virol. 1988;62:1084–1087. doi: 10.1128/jvi.62.3.1084-1087.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataria RS, Tiwari AK, Butchaiah G, Kataria JM, Skinner MA. Sequence analysis of the VP2 gene hypervariable region of infectious bursal disease viruses from India. Avian Pathol. 2001;30:501–507. doi: 10.1080/03079450120078699. [DOI] [PubMed] [Google Scholar]
- 18.Lasher H, Shane S. Infectious bursal disease. World’s Poult Sci J. 1994;50:133–166. doi: 10.1079/WPS19940013. [DOI] [Google Scholar]
- 19.Lazarus D, Pasmanik-Chor M, Gutter B, Gallili G, Barbakov M, Krispel S, Pitcovski J. Attenuation of very virulent infectious bursal disease virus and comparison of full sequences of virulent and attenuated strains. Avian Pathol. 2008;37:151–159. doi: 10.1080/03079450801910206. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Vakharia VN. VP1 protein of infectious bursal disease virus modulates the virulence in vivo. Virology. 2004;330:62–73. doi: 10.1016/j.virol.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Mittal D, Jindal N, Gupta SL, Kataria RS, Singh K, Tiwari AK. Molecular characterization of Indian isolates of infectious bursal disease virus from broiler chickens: full Length Research Paper. DNA Seq. 2006;17:431–439. doi: 10.1080/10425170601017160. [DOI] [PubMed] [Google Scholar]
- 22.Mundt E, Beyer J, Muller H. Identification of a novel viral protein in infectious bursal disease virus-infected cells. J Gen Virol. 1995;76(Pt 2):437–443. doi: 10.1099/0022-1317-76-2-437. [DOI] [PubMed] [Google Scholar]
- 23.Qi X, Gao H, Gao Y, Qin L, Wang Y, Gao L, Wang X. Naturally occurring mutations at residues 253 and 284 in VP2 contribute to the cell tropism and virulence of very virulent infectious bursal disease virus. Antiviral Res. 2009;84:225–233. doi: 10.1016/j.antiviral.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Qi X, Zhang L, Chen Y, Gao L, Wu G, Qin L, Wang Y, Ren X, Gao Y, Gao H. Mutations of residues 249 and 256 in VP2 are involved in the replication and virulence of infectious bursal disease virus. PLoS One. 2013;8:e70982. doi: 10.1371/journal.pone.0070982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudd M, Heine H, Sapats S, Parede L, Ignjatovic J. Characterisation of an Indonesian very virulent strain of infectious bursal disease virus. Arch Virol. 2002;147:1303–1322. doi: 10.1007/s00705-002-0817-3. [DOI] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Sreedevi B, Jackwood D. Real-time reverse transcriptase-polymerase chain reaction detection and sequence analysis of the VP2 hypervariable region of Indian very virulent infectious bursal disease isolates. Avian Dis. 2007;51:750–757. doi: 10.1637/0005-2086(2007)51[750:RRTCRD]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Berg TP, Morales D, Eterradossi N, Rivallan G, Toquin D, Raue R, Zierenberg K, Zhang MF, Zhu YP, Wang CQ, Zheng HJ, Wang X, Chen GC, Lim BL, Muller H. Assessment of genetic, antigenic and pathotypic criteria for the characterization of IBDV strains. Avian Pathol. 2004;33:470–476. doi: 10.1080/03079450400003650. [DOI] [PubMed] [Google Scholar]
- 30.Van Loon A, De Haas N, Zeyda I, Mundt E. Alteration of amino acids in VP2 of very virulent infectious bursal disease virus results in tissue culture adaptation and attenuation in chickens. J Gen Virol. 2002;83:121–129. doi: 10.1099/0022-1317-83-1-121. [DOI] [PubMed] [Google Scholar]
- 31.von Einem UI, Gorbalenya AE, Schirrmeier H, Behrens SE, Letzel T, Mundt E. VP1 of infectious bursal disease virus is an RNA-dependent RNA polymerase. J Gen Virol. 2004;85:2221–2229. doi: 10.1099/vir.0.19772-0. [DOI] [PubMed] [Google Scholar]
- 32.Wei Y, Li J, Zheng J, Xu H, Li L, Yu L. Genetic reassortment of infectious bursal disease virus in nature. Biochem Biophys Res Commun. 2006;350:277–287. doi: 10.1016/j.bbrc.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 33.Wei Y, Yu X, Zheng J, Chu W, Xu H, Yu X, Yu L. Reassortant infectious bursal disease virus isolated in China. Virus Res. 2008;131:279–282. doi: 10.1016/j.virusres.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Yu F, Qi X, Yuwen Y, Wang Y, Gao H, Gao Y, Qin L, Wang X. Molecular characteristics of segment B of seven very virulent infectious bursal disease viruses isolated in China. Virus Genes. 2010;41:246–249. doi: 10.1007/s11262-010-0499-y. [DOI] [PubMed] [Google Scholar]
- 35.Yuwen Y, Gao Y, Gao H, Qi X, Li T, Liu W, Wang X. Sequence analysis of the VP2 hypervariable region of eight very virulent infectious bursal disease virus isolates from the northeast of China. Avian Dis. 2008;52:284–290. doi: 10.1637/8175-111707-Reg.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.