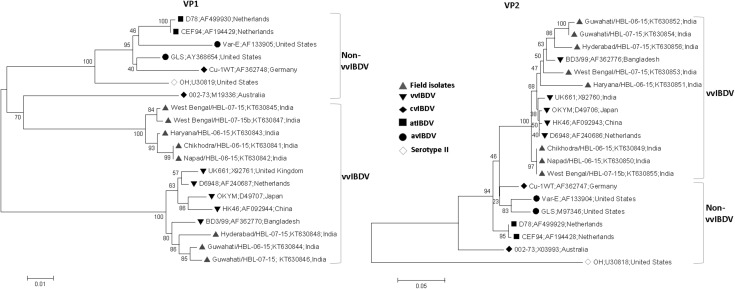
Fig. 3.
Phylogenetic analysis of IBDV field isolates and known IBDV strains based on the VP1 B-marker region and VP2 hypervariable region. Tree was constructed by Neighbor-Joining method using sequences of 8 field strains, and 12 known IBDV strains of different subtypes. Values at the nodes indicates bootstraps probability as determined by 1000 re-sampling. vvIBDV: very virulent IBDV, cvIBDV: classical virulent IBDV, atIBDV: attenuated IBDV, avIBDV: antigenic variant IBDV. Serotype II is a non-pathogenic IBDV strains. Name of the IBDV isolates, their accession numbers and country of origin are given in the order