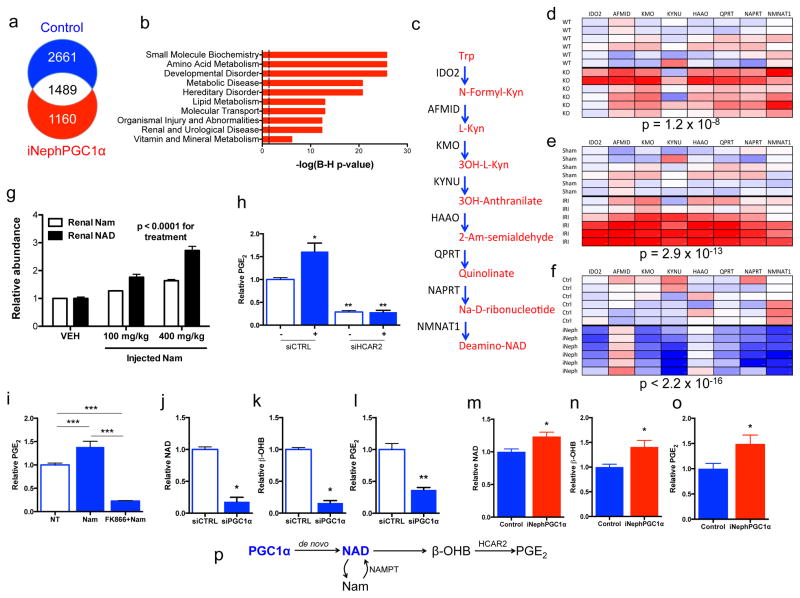
Figure 3. Nam induces β-OHB downstream of PGC1α to augment PGE2.
a, Renal RNA sequencing 24h after IRI or sham operation in controls vs. iNephPGC1α mice with enumerated transcripts. b, Pathway analysis of 1160 transcripts unique to post-ischemic iNephPGC1α mice graphed by −log10[Benjamini-Hochberg-corrected p-value]. Dashed line at p<0.05. c, de novo NAD biosynthetic pathway adapted from KEGG (www.genome.jp/kegg). Trp=tryptophan, Kyn=kynurenine, Am=amino, Na=nicotinate. d–f, Heatmaps (red=lower, blue=higher) of intrarenal expression for de novo pathway from KO vs. WT; 24h after sham vs. IRI; and iNephPGC1α vs. controls (n=6/group). P-values by ANOVA. g, Relative renal Nam and NAD 4h after indicated Nam dose. P-value by ANOVA. h, Conditioned-media-PGE2 of renal tubular cells after HCAR2 knockdown with and without HCAR2 stimulation (+,niacin 10mM, n=6/group). i, PGE2 from renal cells following Nam (1μM for 24h) with and without NAMPT inhibitor FK866 (10nM, n=6/group). j–l, Intracellular NAD, conditioned-media beta-hydroxybutyrate (β-OHB), and conditioned-media PGE2 in PGC1α knockdown cells (n=6/group). m–o, Relative renal NAD, β-OHB, and PGE2 in control vs. iNephPGC1α mice (n=6/group). *p<0.05, **p<0.01, ***p<0.001. p, Renal epithelial PGC1α coordinately upregulates de novo NAD biosynthesis, in the absence of which Nam is utilized through the NAMPT-salvage pathway to generate NAD. Consequently, β-OHB accumulates, which signals HCAR2 to induce PGE2. Error bars SEM.