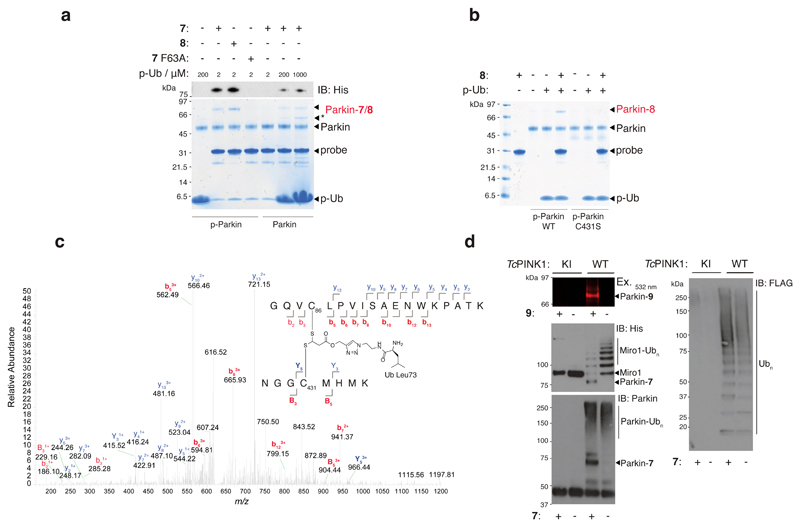
Figure 2. E2~Ub-based probes label the RBR E3 ligase Parkin in an activity-dependant manner.
(a) Coomassie stained reducing SDS-PAGE and anti-His immunoblotting reveals that 7 and 8 (10 μM) form a covalent adduct with p-Parkin (2 μM) in the presence of p-Ub (2 μM) (lanes 2 and 3). Probe 7 F63A (predicted to abolish E3 binding) failed to label Parkin under the same conditions (lane 4). Non-phosphorylated Parkin failed to undergo labelling with probe 7 in the presence of p-Ub (2 μM) (lane 5). Labelling could be effected by the inclusion of molar excess levels of p-Ub (lanes 6 and 7). * Corresponds to contaminating band from p-Ub preparation. (b) Probe 8 does not label p-Parkin C431S in the presence of p-Ub (lane 8 vs. lane 5). All Parkin species and p-Ub were pre-phosphorylated by treatment with PhPINK1. (c) Annotated tryptic MS/MS spectrum for a tryptic 5+ charged precursor ion of the crosslinked peptide derived from labeling of Parkin with 7 (observed m/z = 625.7106; expected m/z = 625.7126) further confirms probe labeling of Parkin C431. (d) In situ probe labelling of reconstituted substrate ubiquitination assays. Parkin and FLAG-Ub in the reactions were phosphorylated by pre-incubation with TcPINK1. Parkin labelling with fluorescent probe 9 and probe 7 was strictly consistent with the Parkin activity readouts of His-SUMO-Miro1 substrate ubiquitination, Parkin autoubiquitination, and free polyubiquitin chain formation. In all cases, activity was strictly dependent on TcPINK1 activity. Consistent results were obtained over 3 replicate experiments.