Table 2. Crystallographic statistics.
Values in parentheses are for the outermost resolution shell.
| Data set | TK2203 | K2PtCl4 | Zn_ano | Cu_ano |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 1.1000 | 1.0700 | 1.2800 | 1.3700 |
| Resolution (Å) | 50–1.41 (1.44–1.41) | 50–2.18 (2.23–2.18) | 50–2.80 (2.90–2.80) | 50–2.80 (2.90–2.80) |
| Total No. of reflections | 546546 | 224928 | 73406 | 74327 |
| No. of unique reflections | 83940 | 23799 | 11030 | 11264 |
| Mosaicity (°) | 0.25–0.61 | 0.21–0.89 | 0.81–1.01 | 0.86–1.04 |
| Multiplicity | 6.5 | 9.5 | 6.7 | 6.6 |
| Completeness (%) | 97.0 (89.7) | 100 (99.6) | 99.8 (99.4) | 99.8 (99.5) |
| 〈I/σ(I)〉 | 23.8 (2.0) | 20.8 (5.8) | 7.7 (2.4) | 6.7 (2.0) |
| R r.i.m. (%) | 8.0 (84.3) | 10.3 (35.3) | 15.2 (50.9) | 17.3 (59.8) |
| Wilson plot B factor (Å2) | 16.3 | 28.0 | 36.1 | 36.0 |
| Space group | C2 | C2 | C2 | C2 |
| Unit-cell parameters | ||||
| a (Å) | 74.4 | 74.4 | 74.3 | 74.2 |
| b (Å) | 42.5 | 42.7 | 42.4 | 42.3 |
| c (Å) | 143.5 | 143.7 | 143.8 | 143.8 |
| β (°) | 94.5 | 94.7 | 94.6 | 94.6 |
| Phasing | ||||
| Mean figure of merit (SIRAS) | 0.39 [20–3 Å] | |||
| Refinement | ||||
| Resolution limits (Å) | 47.69–1.41 | |||
| No. of reflections (F > 0) | 83930 | |||
| No. of reflections used for the R free set | 4233 | |||
| R/R free † (%) | 16.3/18.6 | |||
| R.m.s.d., bond distances (Å) | 0.007 | |||
| R.m.s.d, bond angles (°) | 1.090 | |||
| No. of molecules per asymmetric unit | 2 | |||
| Protein atoms | 4192 | |||
| Ligands | 5 Zn2+, 1 glycerol | |||
| Solvent molecules | 544 waters | |||
| Average B factor (Å2) | ||||
| Protein | 25.8 | |||
| Ligand | 31.1 | |||
| Water | 33.5 | |||
| Ramachandran plot | ||||
| Favoured (%) | 97.5 | |||
| Allowed (%) | 2.5 | |||
| Rotamer outliers (%) | 0.0 | |||
R = 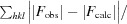
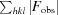 . R
free is the same as R but for a 5% subset of all reflections that were never used in crystallographic refinement.
. R
free is the same as R but for a 5% subset of all reflections that were never used in crystallographic refinement.
