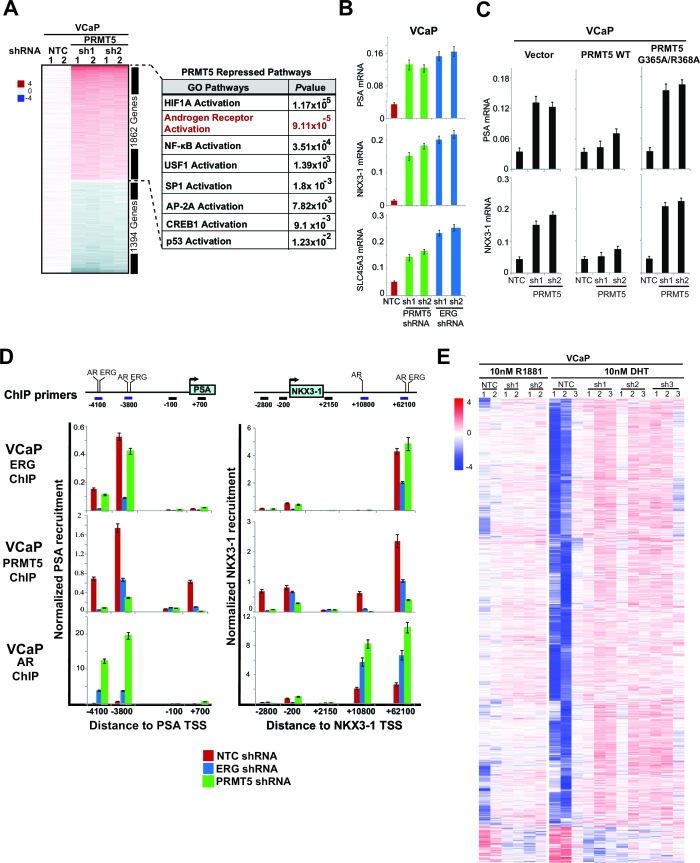
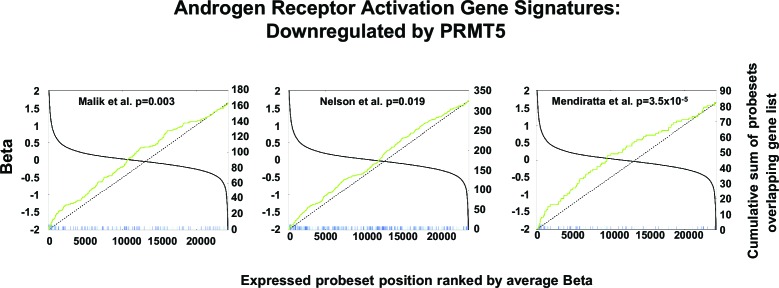
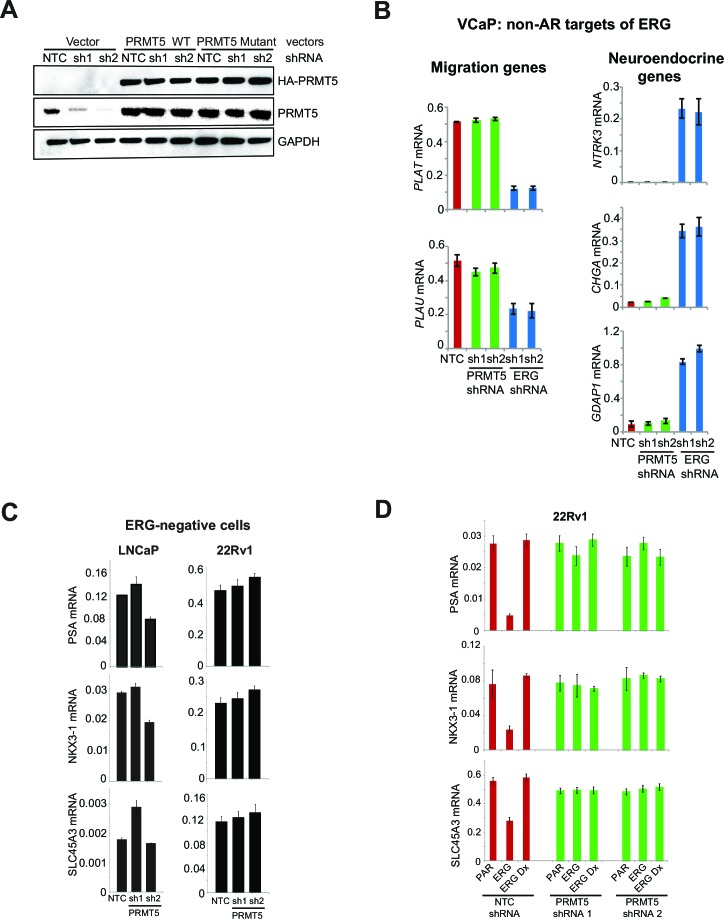
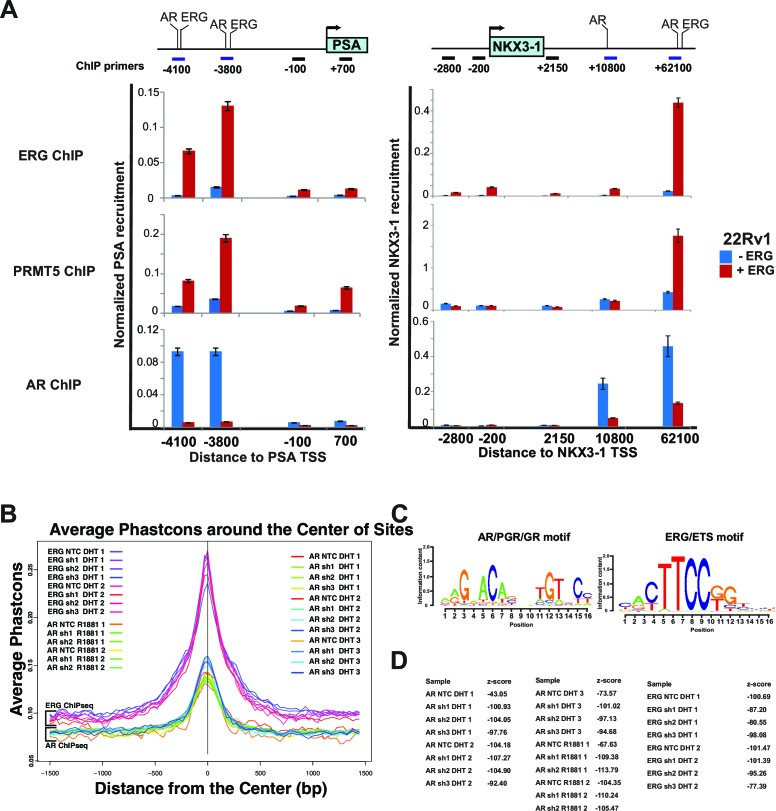
Figure 2. PRMT5 is an ERG-dependent inhibitor of AR signaling.
(A) Heat map showing all genes upregulated (red) or downregulated (blue) by at least 1.5 fold following knockdown with PRMT5 shRNA1 (sh1) or shRNA2 (sh2) compared to NTC shRNA. Rows represent probe sets; columns represent individual samples (technical replicates are marked by 1 or 2). Table indicates pathways significantly upregulated by PRMT5 knockdown (see Materials and methods, and Supplementary file 3 for significantly downregulated pathways). (B) qRT-PCR of AR targets PSA, NKX3-1, and SLC45A3 in VCaP cells expressing the noted shRNA constructs. Expression levels were normalized as described in Materials and methods; bars represent + SEM of three biological replicates, each with three technical repeats. (C) qRT-PCR of PSA and NKX3-1 from VCaP cells expressing the noted shRNA constructs alongside cDNAs expressing vector control (Vector), wild-type (WT) PRMT5, or a catalytically dead PRMT5 mutant (G365A/R368A). Data and error bars represented as in (B). (D) Top panels: cartoons of the PSA and NKX3-1 loci. ERG and AR binding sites (and control regions) are noted and numbered relative to the transcription start site (TSS) as described in Materials and methods. Bottom panels: ERG, PRMT5, and AR ChIP qPCR for the noted regions of PSA (left) or NKX3-1 (right) in VCaP cells upon ERG or PRMT5 knockdown. Normalization to IgG control ChIP is as described in Materials and methods; error bars represent + SEM of three biological replicates, each with three technical repeats. (E) Heatmap visualization of AR binding from ChIP-sequencing data as determined by normalized reads across the AR Cistrome (Materials and methods) in replicate samples induced using AR ligands (DHT or R881 as indicated) and harboring inducible PRMT5 shRNA1 (sh1), shRNA2 (sh2), or shRNA3 (sh3) compared to NTC shRNA. 1659 peaks show differential binding with at least 1.5 fold difference (p-value of 0.01, q-value 0.151). The majority of differentially bound sites exhibit increased binding (6% of the total Cistrome) under PRMT5 knockdown conditions.