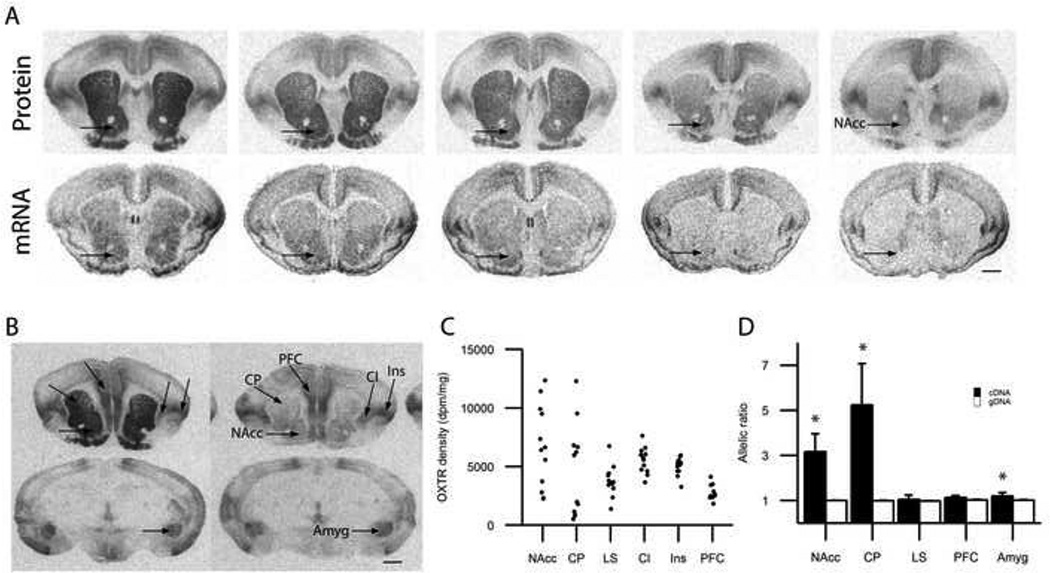
Figure 1.
Region-specific variation in neural expression of the prairie vole Oxtr gene is caused by cis-regulatory elements. (a) An illustration that OXTR protein density (visualized by receptor autoradiography) and Oxtr mRNA levels (visualized by in situ hybridization) vary in the striatum, including the nucleus accumbens (NAcc). (b) In contrast to the individual differences in NAcc and CP OXTR density, other brain regions show less variation. Black scale bar represents 100 µm. (c) Quantification of individual variation in OXTR density. Each dot represents OXTR density (dpm/mg) for an individual prairie vole (N=12, males). NAcc, nucleus accumbens; CP, caudate putamen; LS, dorsal lateral septum; Cl, claustrum; Ins, insular cortex; PFC, prefrontal cortex; Amyg, amygdala. (d) Allelic imbalance was calculated as the average allelic ratio (%T/%C) for cDNA and genomic DNA (gDNA) from animals heterozygous at NT204321 (N=8). A significant allelic imbalance was detected in the cDNA derived from the nucleus accumbens (NAcc), caudate putamen (CP) and amygdala (Amyg), but not in cDNA derived from the prefrontal cortex (PFC) or lateral septum (LS). The striatal regions had very high allelic imbalance, with 3–5 fold differences between alleles. gDNA T and C-alleles are amplified at equal levels in all tissues. * cDNA allelic ratio is significantly greater than a threshold calculated by the mean of the gDNA allelic ratio + 3 gDNA standard deviations. Data are expressed as mean ± standard error of the mean (SEM). Except for CP gDNA (n=6), n=7.