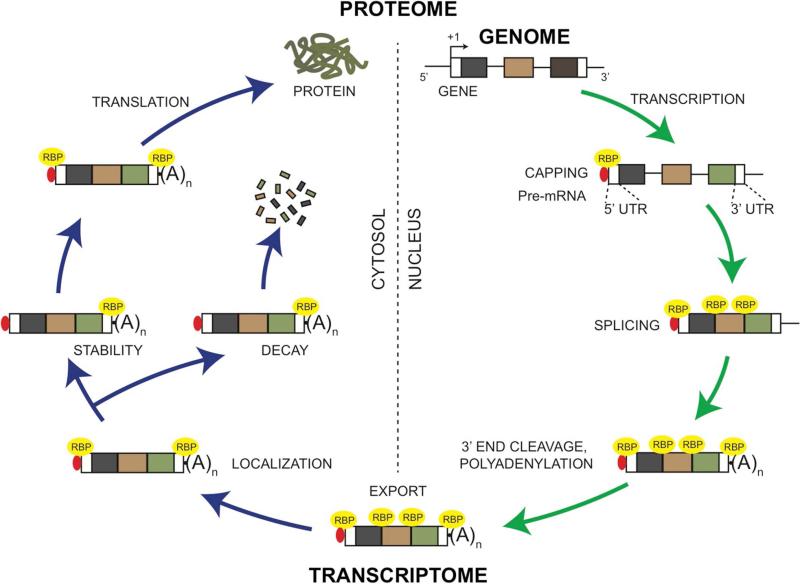
Figure 1. Control of the eukaryotic mRNA by RNA binding proteins.
RNA binding proteins (RBPs) function in distinct regulatory events in the mRNA life-cycle. During transcription of a gene to pre-mRNA, the nascent transcript is capped with 7-methylguanosine to stabilize the mRNA, a process that is facilitated by RBPs such as RAM. RBPs bind to the 5’-Cap to form the Cap binding complex and mediate further control. Excision of the intronic regions from the pre-mRNA can occur co-transcriptionally, a process in which RBPs can bind to the splicing machinery or the exon-intron junctions to drive tissue-specific splicing reactions. The 3’ end of the pre-mRNA is cleaved at a specific site followed by addition of 150-200 adenosine residues (Poly(A) tail) to form a mature mRNA, a process that is facilitated by RBPs such as Poly(A)-binding protein (Pabp). The mature mRNA is then bound by specialized RBPs and exported to the cytosol. In the cytosol, binding of RBPs (e.g. Stau1 or Zbp1) to either the 3’ UTR or the 5’ UTR facilitate the localization mRNA to specific regions for site-specific translation in cells such as neurons or fibroblasts. The localized mRNA is either stabilized or degraded by RBPs binding to sequence-specific sites such as the ARE (AU-rich element) in its 3’UTR. Within the cytosol, RBPs facilitate translation of mRNA into polypeptide. Alternately, mRNA can be recruited to RNA granules such as Processing bodies (P-bodies), Stress granules or other ribonucleoprotein (RNP) complexes for stability, localized translation, silencing or decay (not shown).