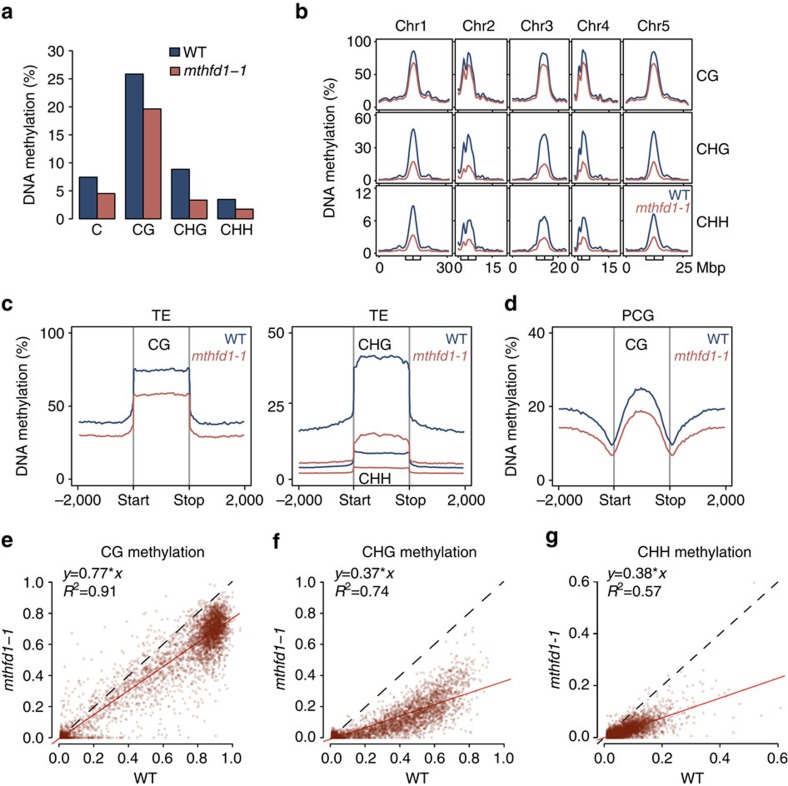
Figure 2. DNA methylation is globally decreased in mthfd1-1 mutants.
(a) Average genome-wide DNA methylation for all Cs and in individual sequence contexts (H=C, A or T). (b) Chromosomal distribution of fractional DNA methylation in individual sequence contexts. Boxes and vertical lines inside boxes mark pericentromeric regions and centromeres, respectively. (c,d) Average distribution of DNA methylation over TEs (c) and PCGs (d) and the flanking 2,000 bp in individual sequence contexts. (e–g) Comparison of DNA methylation levels in 5,000 random 100 bp bins with WT methylation levels >0.01 in CG (e), CHG (f) and CHH (g) contexts. Red line: linear regression between mthfd1-1 and WT levels; corresponding coefficients are shown in top left corners. Dashed: identity line.