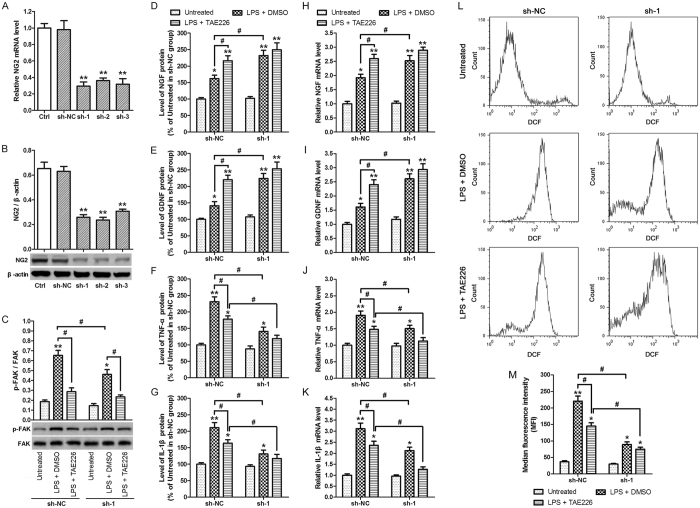
Figure 5. Analysis of NG2 function using NG2 shRNA.
Microglial cells were transfected with control shRNA (sh-NC), NG2 shRNA1 (sh-1), NG2 shRNA2 (sh-2) or NG2 shRNA3 (sh-3) for 48 h. (A) NG2 mRNA expression as determined by qRT-PCR. Data were normalized to GAPDH. **p < 0.01 vs. non-transfected control cells (Ctrl). (B) NG2 protein expression as determined by western blot analysis. Data were normalized to β-actin. **p < 0.01 vs. non-transfected control cells (Ctrl). (C–M) Cells transfected with control shRNA (sh-NC) or NG2 shRNA1 (sh-1) were pretreated with either 10 μM of TAE226 or DMSO for 2 h, and then treated with 10 ng/ml LPS for 6 h. (C) p-FAK and FAK expression as determined by western blotting. The ratios of phosphorylated to total FAK were quantified by densitometric analysis. NGF (D), GDNF (E), TNF-α (F) and IL-1β (G) levels in the culture medium were measured by ELISA. Data were expressed as percentage of levels in untreated of sh-NC group (NGF, 100% = 9.16 ± 1.02 pg/mL; GDNF, 100% = 42.02 ± 5.19 pg/mL; TNF-α, 100% = 60.75 ± 8.31 pg/mL; IL-1β, 100% = 81.29 ± 10.57 pg/mL). NGF (H), GDNF (I), TNF-α (J) and IL-1β (K) mRNA expression of microglial cells as determined by qRT-PCR. Data were normalized to GAPDH. (L) Intracellular ROS levels determined by flow cytometry using DCFH-DA staining. (M) The intracellular ROS data were quantified by the median fluorescence intensity (MFI). *p < 0.05, **p < 0.01 vs. untreated group. #p < 0.05. Data are represented as the mean ± s.d. (n = 3, technical replicates). Each experiment was repeated on at least two separate occasions.