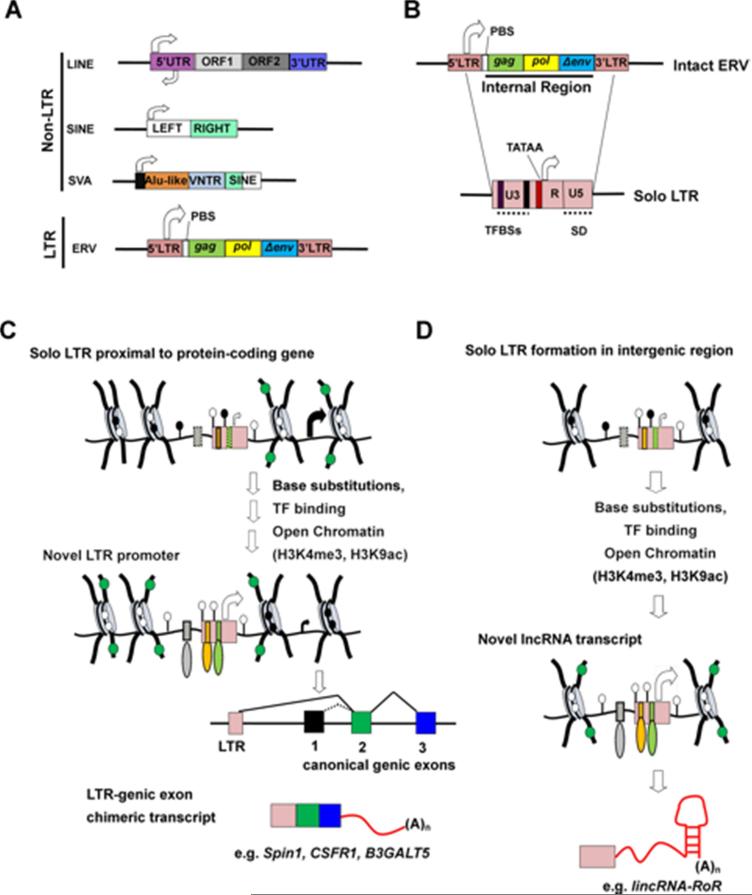
Figure 1. Structure of an intact ERV and solo LTR and the molecular mechanisms of LTR exaptation as protein-coding or lncRNA promoters.
(A) Schematic of non-LTR retrotransposons which include SINEs (i.e. Alus), LINEs (i.e. L1Hs), and SVAs (in humans) and LTR retrotransposons, which include many lineage/species-specific subfamilies. Most LINE elements are truncated at the 5’ end, thus lacking the 5’UTR promoter and TSS. (B) Full-lengthERVs have 5’ and 3’ LTRs, and an “internal” region that includes a primer-binding site (PBS) involved in priming reverse transcription, and retroviral ORFs gag, pol and a truncated or mutated env gene (Δenv). Recombination between 5’ and 3’ LTRs deletes the internal region, generating ‘solo’ LTRs (not to scale), which consist of unique 3’ (U3) and 5’ (U5) regions and a regulatory region (R) containing the TSS (white arrow). LTRs often harbour different combinations of TFBSs (green and orange rectangles) in addition to core Pol II promoter elements (such as TATA box shown in red) and may also contain a splice donor (SD) site (dashed line) within the U5 region. (C) LTR exaptation as a protein-coding gene promoter. In a developmental/tissue specific context, particularly in cell types undergoing epigenetic reprogramming (e.g. early embryo, placenta or germline) a hypomethylated solo LTR (pink rectangle) 5’ of a protein-coding gene (or in an intragenic region) may become exapted as a novel promoter (black circles represent DNA methylation). The process may involve base substitutions in a neighbouring near-consensus TFBS (grey rectangle with dash outline) and a near-consensus site within the LTR (green rectangle with dash outline), which then form a positive genetic interaction with another LTR-derived TFBS (orange rectangle) a mechanism termed ‘epistatic capture’ (Emera and Wagner, 2012a). This leads to synergy in the binding of several TFs (grey, orange and green ovals), deposition of “active” histone modifications, such as H3K4me3 and H3K9ac (green circles) and robust transcription initiation from the LTR-derived promoter. The canonical genic promoter may be DNA methylated as a consequence of such transcription. This process generates LTR-genic exon chimeric transcripts, where exon 1 is derived from the LTR and splicing occurs from the internal LTR SD (or from a cryptic SD site in the intervening genomic sequence downstream of the active LTR) to the first downstream exon with a splice-acceptor site, generally exon 2. Examples of such chimeric transcripts include Spin1 in mouse and CSFR1 and B3GALT5 in human. Arrow sizes indicate relative level of transcription from each promoter. (D) LTR exaptation as a promoter for a novel lncRNA. Through a process as in (C), a newly formed intergenic solo LTR without an SD site could initiate de novo lncRNA transcription, forming a novel lncRNA gene. An example of this is the lincRNA-RoR transcript.