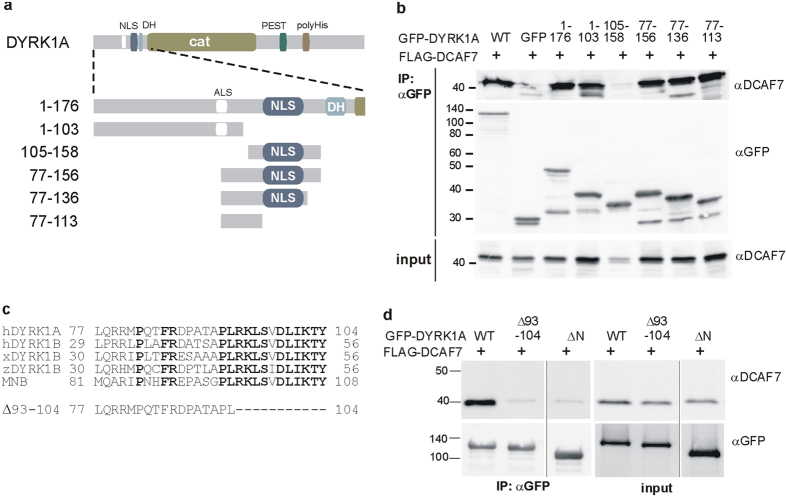
Figure 2. Mapping of the DCAF7-interacting sequence in DYRK1A.
(a) Schematic representation of DYRK1A domain structure and deletion clones. - Deletion constructs tested for DCAF7 binding are illustrated in a close-up of the N-terminal domain. ALS, alternatively spliced segment of 9 amino acids that is absent in the short splicing variant; NLS, nuclear localization sequence; DH, DYRK homology box; cat, catalytic domain; PEST, proline-, glutamic acid-, serine-, threonine-rich region; polyHis, poly-histidine tract. (b) Co-IP of FLAG-DCAF7 with GFP-DYRK1A deletion constructs. - HeLa cells co-expressing FLAG-DCAF7 and the indicated GFP-DYRK1A deletion constructs or unfused GFP were used for GFP-IP. Binding of DCAF7 was analysed by immunoblotting. (c) Alignment of the minimal DCAF7 binding region in vertebrate class 1 DYRKs and the Drosophila ortholog MNB. - A deletion was introduced in the full length kinase to narrow down the binding region (Δ93–104). (d) FLAG-DCAF7 does not bind to GFP-DYRK1A-Δ93–104. – Binding of FLAG-DCAF7 to GFP-DYRK1A constructs was assayed by co-IP as in panel (b). GFP-DYRK1A-ΔN lacks the N-terminal domain (Δ1–134) and was used as a negative control.