The approximately planar (E,E)-2′,4′-dihydroxyacetophenone azine molecule is located on an inversion centre and linked with dimethylformamide solvent molecules via O—H⋯O hydrogen bonds.
Keywords: crystal structure; hydrogen bond; (E,E)-2′,4′-dihydroxyacetophenone azine
Abstract
In the title compound {systematic name: 4,4′-[1,1′-(hydrazinediylidene)bis(ethan-1-yl-1-ylidene)]bis(benzene-1,3-diol)}, C16H16N2O4·2C3H7NO, the (E,E)-2′,4′-dihydroxyacetophenone azine molecule is centrosymmetric, the mid-point of the N—N bond being located on an inversion centre. All the non-H atoms of the azine molecule are approximately coplanar, the maximum deviation being 0.017 (2) Å. An intramolecular O—H⋯N hydrogen bond occurs between the azine N atom and the hydroxy group. In the crystal, azine and dimethylformamide solvent molecules are linked by O—H⋯O hydrogen bonds.
Chemical context
Hydrazones are important compounds due to their possible applications in material and coordination chemistry. Fluorescence properties of hydrazones have been reported (Qin et al., 2009 ▸). Many organometallic compounds containing acylhydrazone ligands have also been synthesized for their potential magneto-chemical properties (Guo et al., 2010 ▸). In particular, they have received increasing interest for their biological activity as antioxidants (Kitaev et al., 1970 ▸), and their antimicrobial (Ramamohan et al., 1995 ▸) and antiviral properties (El-Tabl et al., 2008 ▸; Rollas & Küçükgüzel, 2007 ▸).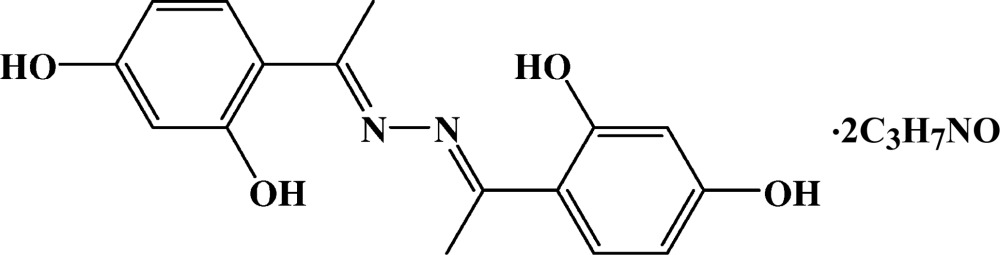
Although 2′,4′-dihydroxyacetophenone azine has been prepared and studied as a fluorescent probe, its structure has not been reported. As a part of our studies on synthesis and structural peculiarities of Schiff base ligands derived from 2′,4′-dihydroxyacetophenone and hydrazine, we determined the structure of the title compound, (E,E)-2′,4′-dihydroxyacetophenone azine dimethylformamide disolvate, (I).
Structural commentary
The molecular structure of the title compound is depicted in Fig. 1 ▸. The asymmetric unit contains one half-molecule of (E,E)-2′,4′-dihydroxyacetophenone azine and one dimethylformamide (DMF) molecule. The complete azine molecule is centrosymmetric and exists in an E,E configuration with respect to the two C=N bonds. The N1—C2 bond length of 1.301 (3) Å shows double-bond character. The C—O bond lengths [1.349 (3) and 1.358 (3) Å] are comparable with similar bonds in related structures (Chantrapromma et al., 2011 ▸; Tai et al., 2008 ▸). All the non-H atoms of the azine molecule are approximately coplanar. The nine atoms (i.e. N1, C1 and C2, and the six C atoms in the benzene ring) are essentially planar, with a mean deviation of 0.0024 Å. Each hydroxy group is nearly coplanar with its attached benzene ring; the r.m.s. deviation is 0.0045 Å for the seven non-H atoms. Intramolecular O—H⋯N hydrogen bonds exist in the azine molecule (Table 1 ▸).
Figure 1.
The molecular structure of the title compound, showing the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level. Only one DMF solvent molecule is shown. [Symmetry code: (i) −x + 1, −y + 1, −z + 1.]
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 0.82 | 1.82 | 2.543 (2) | 147 |
| O2—H2⋯O3i | 0.82 | 1.84 | 2.649 (3) | 171 |
Symmetry code: (i)  .
.
Supramolecular features
In the crystal of (I), intermolecular O—H⋯O hydrogen bonds exist between azine molecules and DMF molecules (Table 1 ▸ and Fig. 2 ▸).
Figure 2.
The crystal packing of the title compound. Hydrogen bonds are shown as dashed lines. [Symmetry codes: (i) −x + 1, −y + 1, −z + 1; (ii) x − 1, y + 1, z.]
Database survey
A search of Cambridge Structural Database (Groom & Allen, 2014 ▸) for acetophenone azine gave 105 hits (excluding organometallics). There are four reported crystal structures of acetophenone azine containing hydroxy groups at the 2-position of benzene rings: (E,E)-2,2′-(1,1′-azinodiethylidyne)diphenol (Tai et al., 2008 ▸), (E,E)-4,4′-dichloro-2,2′-(1,1′-azinodiethylidyne)diphenol (Chang et al., 2007 ▸), (E,E)-3,3′-diethoxy-2,2′-(1,1′-azinodiethylidyne)diphenol (Fayos et al., 1980 ▸) and (E,E)-4,4′-dimethoxy-2,2′-(1,1′-azinodiethylidyne)diphenol (Zhang et al., 2008 ▸).
Synthesis and crystallization
A mixture of 2′,4′-dihydroxyacetophenone (3.06 g, 20 mmol), hydrazine sulfate (1.28 g, 10 mmol) and triethylamine (3.03 g, 30 mmol) in ethanol (40 ml) was heated under reflux for 24 h. After cooling, the precipitate was filtrated and washed with water to afford a yellow solid. Crystals of the title compound suitable for X-ray diffraction were obtained by slow evaporation of a solution of the solid in DMF at room temperature for 5 d (yield 1.20 g, 75%; m.p: 484–485 K). 1H NMR (300 MHz, CDCl3): δ 13.59 (s, 2H, OH), 10.14 (s, 2H, OH), 7.58–7.61 (d, 2H, ArH), 6.37–6.41 (d, 2H, ArH), 6.30–6.31 (s, 2H, ArH), 3.34 (d, 6H, CH3).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. H atoms were placed geometrically (C—H = 0.93–0.96 Å and O—H = 0.82 Å) and refined as riding, with U iso(H) = 1.2U eq(C) for aromatic H atoms or 1.5U eq(C,O) for methyl and hydroxy groups.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C16H16N2O4·2C3H7NO |
| M r | 446.50 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 298 |
| a, b, c (Å) | 6.1616 (7), 7.3109 (8), 13.4537 (15) |
| α, β, γ (°) | 96.771 (1), 103.049 (2), 96.607 (1) |
| V (Å3) | 579.96 (11) |
| Z | 1 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.48 × 0.43 × 0.21 |
| Data collection | |
| Diffractometer | Bruker SMART CCD area-detector |
| Absorption correction | Multi-scan (SADABS; Sheldrick, 1996 ▸) |
| T min, T max | 0.956, 0.981 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 2902, 2001, 1313 |
| R int | 0.026 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.055, 0.187, 1.02 |
| No. of reflections | 2001 |
| No. of parameters | 149 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.25 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989016003686/xu5885sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016003686/xu5885Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016003686/xu5885Isup3.cml
CCDC reference: 1457201
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Financial support from the Natural Science Foundation of Shanxi Province (No. 2013011011-4) is gratefully acknowledged.
supplementary crystallographic information
Crystal data
| C16H16N2O4·2C3H7NO | Z = 1 |
| Mr = 446.50 | F(000) = 238 |
| Triclinic, P1 | Dx = 1.278 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.1616 (7) Å | Cell parameters from 1119 reflections |
| b = 7.3109 (8) Å | θ = 3.0–26.5° |
| c = 13.4537 (15) Å | µ = 0.09 mm−1 |
| α = 96.771 (1)° | T = 298 K |
| β = 103.049 (2)° | Block, colorless |
| γ = 96.607 (1)° | 0.48 × 0.43 × 0.21 mm |
| V = 579.96 (11) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2001 independent reflections |
| Radiation source: fine-focus sealed tube | 1313 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.026 |
| phi and ω scans | θmax = 25.0°, θmin = 3.0° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −6→7 |
| Tmin = 0.956, Tmax = 0.981 | k = −8→7 |
| 2902 measured reflections | l = −15→15 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.055 | H-atom parameters constrained |
| wR(F2) = 0.187 | w = 1/[σ2(Fo2) + (0.1079P)2 + 0.0859P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 2001 reflections | Δρmax = 0.28 e Å−3 |
| 149 parameters | Δρmin = −0.25 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.17 (2) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.4279 (3) | 0.5237 (2) | 0.53085 (13) | 0.0391 (5) | |
| N2 | 0.5727 (4) | 0.1587 (3) | 0.91556 (16) | 0.0566 (6) | |
| O1 | 0.2575 (3) | 0.7659 (2) | 0.62846 (12) | 0.0537 (5) | |
| H1 | 0.3372 | 0.7270 | 0.5919 | 0.081* | |
| O2 | −0.2843 (3) | 0.5582 (3) | 0.80524 (14) | 0.0679 (6) | |
| H2 | −0.2793 | 0.6712 | 0.8190 | 0.102* | |
| O3 | 0.7469 (4) | −0.0813 (3) | 0.87057 (18) | 0.0899 (8) | |
| C1 | 0.3219 (5) | 0.1857 (3) | 0.5203 (2) | 0.0574 (7) | |
| H1A | 0.2229 | 0.1462 | 0.4531 | 0.086* | |
| H1B | 0.2779 | 0.1098 | 0.5681 | 0.086* | |
| H1C | 0.4737 | 0.1732 | 0.5172 | 0.086* | |
| C2 | 0.3080 (3) | 0.3858 (3) | 0.55569 (15) | 0.0374 (6) | |
| C3 | 0.1542 (3) | 0.4334 (3) | 0.62014 (15) | 0.0368 (6) | |
| C4 | 0.1368 (4) | 0.6208 (3) | 0.65420 (16) | 0.0403 (6) | |
| C5 | −0.0090 (4) | 0.6624 (3) | 0.71594 (16) | 0.0458 (6) | |
| H5 | −0.0180 | 0.7861 | 0.7380 | 0.055* | |
| C6 | −0.1404 (4) | 0.5221 (4) | 0.74482 (17) | 0.0479 (6) | |
| C7 | −0.1270 (4) | 0.3382 (4) | 0.71238 (18) | 0.0528 (7) | |
| H7 | −0.2153 | 0.2431 | 0.7316 | 0.063* | |
| C8 | 0.0176 (4) | 0.2968 (3) | 0.65154 (17) | 0.0472 (6) | |
| H8 | 0.0249 | 0.1723 | 0.6304 | 0.057* | |
| C9 | 0.5795 (5) | −0.0016 (5) | 0.8617 (2) | 0.0696 (9) | |
| H9 | 0.4489 | −0.0589 | 0.8136 | 0.084* | |
| C10 | 0.7669 (5) | 0.2551 (5) | 0.9909 (2) | 0.0808 (9) | |
| H10A | 0.8729 | 0.1706 | 1.0084 | 0.121* | |
| H10B | 0.7228 | 0.3039 | 1.0516 | 0.121* | |
| H10C | 0.8355 | 0.3556 | 0.9632 | 0.121* | |
| C11 | 0.3699 (5) | 0.2460 (5) | 0.8988 (3) | 0.0889 (10) | |
| H11A | 0.2480 | 0.1611 | 0.8530 | 0.133* | |
| H11B | 0.3951 | 0.3568 | 0.8687 | 0.133* | |
| H11C | 0.3324 | 0.2778 | 0.9635 | 0.133* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0427 (11) | 0.0333 (11) | 0.0416 (10) | 0.0063 (8) | 0.0117 (8) | 0.0035 (8) |
| N2 | 0.0545 (13) | 0.0620 (15) | 0.0553 (12) | 0.0128 (11) | 0.0150 (10) | 0.0088 (11) |
| O1 | 0.0704 (11) | 0.0331 (10) | 0.0647 (11) | 0.0015 (8) | 0.0367 (9) | 0.0024 (7) |
| O2 | 0.0738 (13) | 0.0718 (14) | 0.0713 (12) | 0.0124 (10) | 0.0428 (10) | 0.0122 (10) |
| O3 | 0.0897 (16) | 0.0827 (17) | 0.1017 (17) | 0.0266 (14) | 0.0354 (13) | −0.0059 (13) |
| C1 | 0.0708 (17) | 0.0345 (14) | 0.0745 (17) | 0.0076 (12) | 0.0332 (14) | 0.0080 (12) |
| C2 | 0.0380 (12) | 0.0337 (12) | 0.0377 (11) | 0.0037 (9) | 0.0037 (9) | 0.0065 (9) |
| C3 | 0.0383 (12) | 0.0346 (12) | 0.0351 (11) | 0.0028 (9) | 0.0048 (9) | 0.0057 (9) |
| C4 | 0.0450 (13) | 0.0378 (13) | 0.0364 (11) | 0.0024 (10) | 0.0078 (10) | 0.0064 (9) |
| C5 | 0.0532 (14) | 0.0414 (14) | 0.0427 (12) | 0.0066 (11) | 0.0136 (11) | 0.0020 (10) |
| C6 | 0.0451 (13) | 0.0583 (16) | 0.0411 (12) | 0.0065 (11) | 0.0120 (10) | 0.0081 (11) |
| C7 | 0.0548 (15) | 0.0502 (16) | 0.0561 (15) | −0.0022 (12) | 0.0199 (12) | 0.0155 (11) |
| C8 | 0.0539 (14) | 0.0385 (14) | 0.0497 (13) | 0.0031 (11) | 0.0137 (11) | 0.0102 (10) |
| C9 | 0.0675 (19) | 0.081 (2) | 0.0575 (16) | −0.0021 (16) | 0.0183 (14) | 0.0061 (15) |
| C10 | 0.078 (2) | 0.075 (2) | 0.078 (2) | 0.0069 (17) | 0.0043 (17) | 0.0000 (16) |
| C11 | 0.069 (2) | 0.102 (3) | 0.102 (2) | 0.0268 (19) | 0.0191 (18) | 0.032 (2) |
Geometric parameters (Å, º)
| N1—C2 | 1.301 (3) | C3—C4 | 1.417 (3) |
| N1—N1i | 1.391 (3) | C4—C5 | 1.389 (3) |
| N2—C9 | 1.313 (4) | C5—C6 | 1.380 (3) |
| N2—C10 | 1.430 (4) | C5—H5 | 0.9300 |
| N2—C11 | 1.453 (3) | C6—C7 | 1.381 (3) |
| O1—C4 | 1.349 (3) | C7—C8 | 1.374 (3) |
| O1—H1 | 0.8200 | C7—H7 | 0.9300 |
| O2—C6 | 1.358 (3) | C8—H8 | 0.9300 |
| O2—H2 | 0.8200 | C9—H9 | 0.9300 |
| O3—C9 | 1.232 (3) | C10—H10A | 0.9600 |
| C1—C2 | 1.503 (3) | C10—H10B | 0.9600 |
| C1—H1A | 0.9600 | C10—H10C | 0.9600 |
| C1—H1B | 0.9600 | C11—H11A | 0.9600 |
| C1—H1C | 0.9600 | C11—H11B | 0.9600 |
| C2—C3 | 1.465 (3) | C11—H11C | 0.9600 |
| C3—C8 | 1.396 (3) | ||
| C2—N1—N1i | 116.3 (2) | O2—C6—C5 | 122.1 (2) |
| C9—N2—C10 | 120.9 (2) | O2—C6—C7 | 118.0 (2) |
| C9—N2—C11 | 121.2 (3) | C5—C6—C7 | 119.9 (2) |
| C10—N2—C11 | 117.9 (3) | C8—C7—C6 | 119.6 (2) |
| C4—O1—H1 | 109.5 | C8—C7—H7 | 120.2 |
| C6—O2—H2 | 109.5 | C6—C7—H7 | 120.2 |
| C2—C1—H1A | 109.5 | C7—C8—C3 | 122.9 (2) |
| C2—C1—H1B | 109.5 | C7—C8—H8 | 118.6 |
| H1A—C1—H1B | 109.5 | C3—C8—H8 | 118.6 |
| C2—C1—H1C | 109.5 | O3—C9—N2 | 124.5 (3) |
| H1A—C1—H1C | 109.5 | O3—C9—H9 | 117.8 |
| H1B—C1—H1C | 109.5 | N2—C9—H9 | 117.8 |
| N1—C2—C3 | 116.96 (19) | N2—C10—H10A | 109.5 |
| N1—C2—C1 | 122.62 (19) | N2—C10—H10B | 109.5 |
| C3—C2—C1 | 120.4 (2) | H10A—C10—H10B | 109.5 |
| C8—C3—C4 | 116.5 (2) | N2—C10—H10C | 109.5 |
| C8—C3—C2 | 121.9 (2) | H10A—C10—H10C | 109.5 |
| C4—C3—C2 | 121.6 (2) | H10B—C10—H10C | 109.5 |
| O1—C4—C5 | 117.0 (2) | N2—C11—H11A | 109.5 |
| O1—C4—C3 | 122.42 (19) | N2—C11—H11B | 109.5 |
| C5—C4—C3 | 120.6 (2) | H11A—C11—H11B | 109.5 |
| C6—C5—C4 | 120.7 (2) | N2—C11—H11C | 109.5 |
| C6—C5—H5 | 119.7 | H11A—C11—H11C | 109.5 |
| C4—C5—H5 | 119.7 | H11B—C11—H11C | 109.5 |
| N1i—N1—C2—C3 | −179.63 (19) | C3—C4—C5—C6 | −0.3 (3) |
| N1i—N1—C2—C1 | −0.1 (3) | C4—C5—C6—O2 | 179.9 (2) |
| N1—C2—C3—C8 | −179.94 (18) | C4—C5—C6—C7 | 0.2 (3) |
| C1—C2—C3—C8 | 0.5 (3) | O2—C6—C7—C8 | −179.7 (2) |
| N1—C2—C3—C4 | −0.2 (3) | C5—C6—C7—C8 | 0.0 (4) |
| C1—C2—C3—C4 | −179.69 (19) | C6—C7—C8—C3 | −0.1 (4) |
| C8—C3—C4—O1 | −179.28 (19) | C4—C3—C8—C7 | −0.1 (3) |
| C2—C3—C4—O1 | 0.9 (3) | C2—C3—C8—C7 | 179.7 (2) |
| C8—C3—C4—C5 | 0.3 (3) | C10—N2—C9—O3 | 0.1 (4) |
| C2—C3—C4—C5 | −179.47 (19) | C11—N2—C9—O3 | 178.5 (3) |
| O1—C4—C5—C6 | 179.3 (2) |
Symmetry code: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.82 | 1.82 | 2.543 (2) | 147 |
| O2—H2···O3ii | 0.82 | 1.84 | 2.649 (3) | 171 |
Symmetry code: (ii) x−1, y+1, z.
References
- Bruker (2007). SMART and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chang, J.-G., He, G.-F. & Li, Y.-F. (2007). Acta Cryst. E63, o3982.
- Chantrapromma, S., Jansrisewangwong, P., Chanawanno, K. & Fun, H.-K. (2011). Acta Cryst. E67, o2221–o2222. [DOI] [PMC free article] [PubMed]
- El-Tabl, A. S., El-Saied, F. A., Plass, W. & Al-Hakimi, A. N. (2008). Spectrochim. Acta A Mol. Biomol. Spectrosc. 71, 90–99. [DOI] [PubMed]
- Fayos, J., Martínez-Ripoll, M., García-Mina, M. C., Gonzalez-Martínez, J. & Arrese, F. (1980). Acta Cryst. B36, 1952–1953.
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Guo, Y.-N., Xu, G.-F., Gamez, P., Zhao, L., Lin, S.-Y., Deng, R., Tang, J.-K. & Zhang, H.-J. (2010). J. Am. Chem. Soc. 132, 8538–8539. [DOI] [PubMed]
- Kitaev, Y. P., Buzykin, B. I. & Troepol’skaya, T. V. (1970). Russ. Chem. Rev. 39, 441–456.
- Qin, D.-D., Yang, Z.-Y. & Qi, G.-F. (2009). Spectrochim. Acta A Mol. Biomol. Spectrosc. 74, 415–420. [DOI] [PubMed]
- Ramamohan, L., Shikkargol, R. K., Angadi, S. D. & Kulkarni, V. H. (1995). Asian J. Pure Appl. Chem. 1, 86–89.
- Rollas, S. & Küçükgüzel, Ş. G. (2007). Molecules, 12, 1910–1939. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (1996). SADABS. University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tai, X.-S., Xu, J., Feng, Y.-M. & Liang, Z.-P. (2008). Acta Cryst. E64, o905. [DOI] [PMC free article] [PubMed]
- Zhang, J.-H., Dong, W.-L., Ge, Y.-Q. & Zhao, B.-X. (2008). Acta Cryst. E64, o166. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989016003686/xu5885sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016003686/xu5885Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016003686/xu5885Isup3.cml
CCDC reference: 1457201
Additional supporting information: crystallographic information; 3D view; checkCIF report




