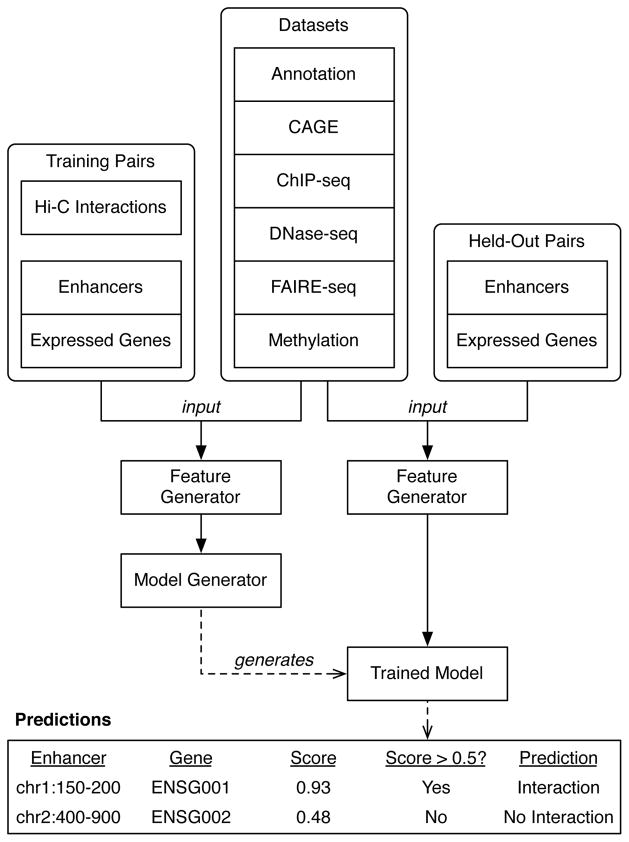
Figure 4.
The TargetFinder pipeline. Features are generated from hundreds of diverse datasets for pairs of enhancers and promoters of expressed genes found to have significant Hi-C interactions (positives), as well as random pairs of enhancers and promoters without significant interactions (negatives). These labeled samples are used to train an ensemble classifier that predicts whether enhancer-promoter pairs from new or held-out samples interact, as well as estimate the importance of each feature for accurate prediction. Classifier predictions are probabilities, and a decision threshold (commonly 0.5 but may be adjusted) converts these to positive or negative prediction labels. This figure excludes selection of minimal predictor sets and evaluation of the accuracy of output predictions using held-out Hi-C interaction data.