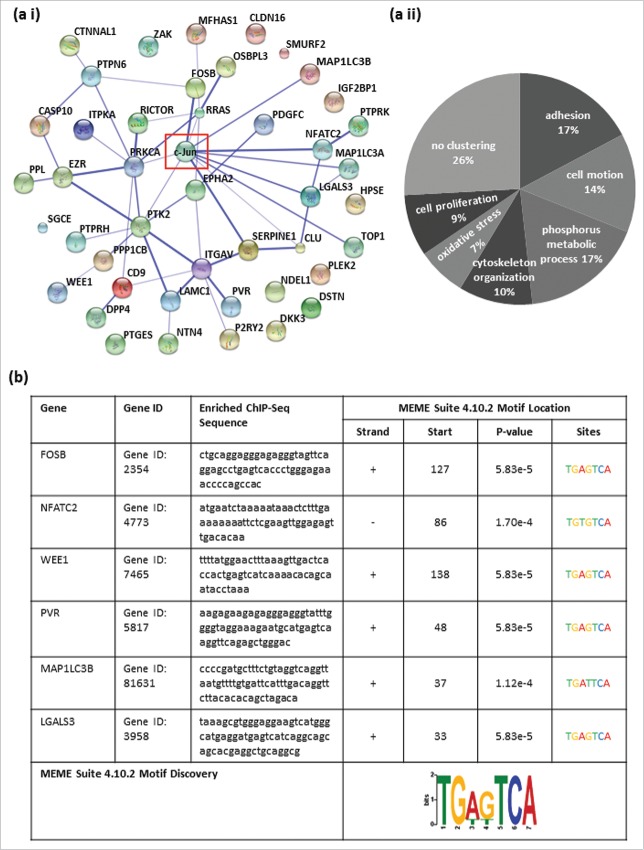
Figure 1.
All identified potential c-Jun target genes share cancer-relevant molecular functions and contain classical or non-classical AP-1 recognition sequences in their promoter/enhancer regions. (a i) Predicted relation between the 44 identified target genes and the transcription factor c-Jun via STRING. (a ii) Functional annotation clustering using the category GOTERM_BP_FAT resulted in 7 different significantly enriched annotation clusters (ACs). The ACs contained genes that could be summarized according to their molecular function: adhesion (enrichment score (ES): 2.76), cell motion (ES: 2.42), phosphorus metabolic process (ES: 1.84), cytoskeleton organization (ES: 1.62), response to oxidative stress (ES: 1.46) and positive regulation of cell proliferation (ES: 1.14). (b) The promoter/enhancer regions of FosB, WEE1, PVR and LGALS3 contained the classical AP-1 binding sequence with 100% agreement (FosB: 5′-TGA G TCA-3′, WEE1: 5′-TGA(C/G)TCA-3′, PVR: 5′-TGAGTCA-3′, and LGALS3: 5′-TGA G TCA-3′). In the promoter/enhancer regions of NFATC2 and MAP1LC3B, the AP-1 binding motifs each contained a one base pair exchange (NFATC2: 5′-TGA C ACA-3′ and MAP1LC3B: 5′-TGA T TCA-3′). The potential c-Jun binding sites were validated in all 6 detected potential target genes as DNA-motifs, identical or similar to the AP-1 recognition sequence, using the Motiv Discovery program MEME.