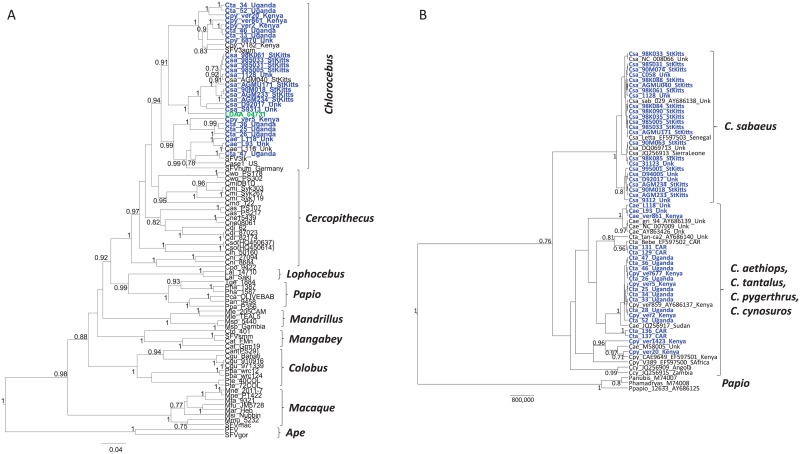
Fig 3. Inference of the evolutionary history of human infection with simian foamy virus (SFV).
A. Bayesian SFV polymerase (pol) maximum clade credibility tree inferred using an alignment of 375-bp and 88 Old World monkey and ape taxa, including 26 new African green monkey sequences (in blue text). The SFV pol sequence from the Côte d’Ivoire patient, LDAA-04731, is shown in green text. B. Chlorocebus mitochondrial DNA phylogenetic relationships inferred by Bayesian analysis of an alignment of 496-bp from 65 taxa. New sequences generated in our study are highlighted in blue text. Animal country of origin given at end of taxa names; CAR = Central African Republic, Unk = provenance unknown. Csa, Chl. sabaeus, Cae, Chl. aethiops, Cta, Chl. tantalus, Cpy, Chl. pygerythrus, Ccy, Chl. cynosuros. Posterior probabilities > 0.7 are given at major nodes in the tree. Scale bar is in units of time.