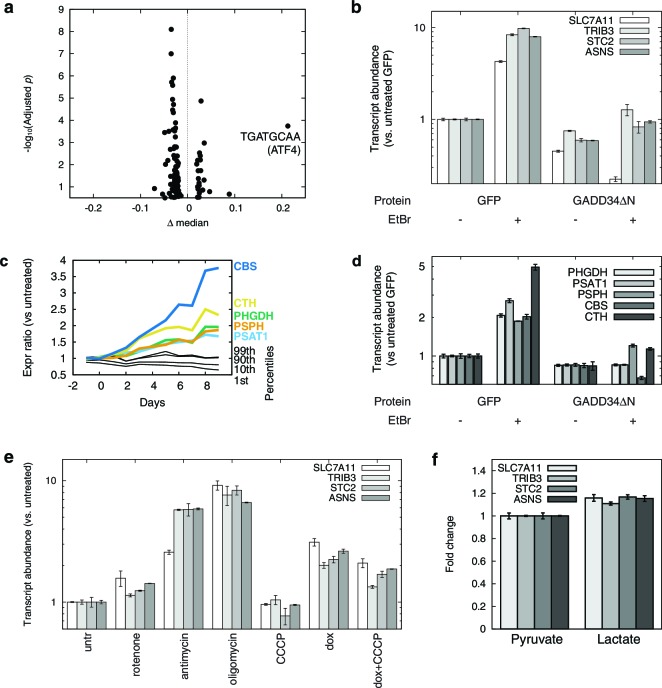
Figure 2. mtDNA depletion activates ATF4.
(a) Volcano plot of motifADE analysis results (see Results). Δ median denotes the normalized rank of the median gene associated with each motif. (b) Fold changes of four of the most-upregulated genes in microarray data, in response to mtDNA depletion by ethidium bromide (EtBr) treatment (100 ng/ml, 9 d), and with expression of either GFP or GADD34ΔN. (c) Activation of serine and homocysteine biosynthesis genes, compared to that of genes at the 1st, 10th, 90th, and 99th percentiles in each transcriptional profiling timepoint. (d) Activation of serine and cysteine biosynthesis genes in response to mtDNA depletion, with and without GADD34ΔN expression. (e) Activation of ATF4 target genes in response to mitochondrial inhibitors. (f) Activation of ATF4 target genes in response to cytoplasmic redox imbanace elicited by lactate. n = 3 for b, d, e, and f.