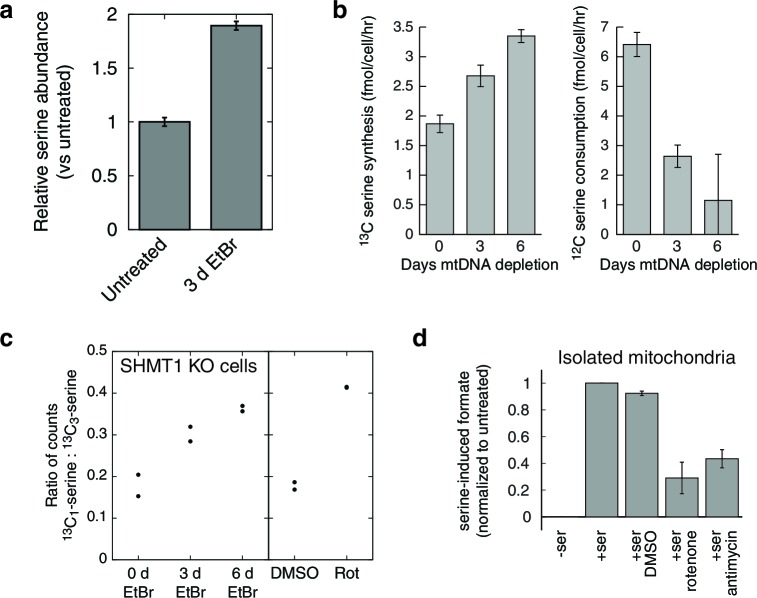
Figure 4. Respiratory chain dysfunction impairs mitochondrial 1C metabolism.
(a) Confirmation of altered serine levels in spent media with EtBr-induced mtDNA depletion. n = 3. (b) Tracing serine metabolism using 13C-labeled glucose. Serine synthesis rates are inferred by labeling cells for 30 min with 13C6-glucose and measuring the amount of 13C3-serine that emerges. Simultaneously, serine consumption rates can be inferred by the amount of unlabeled serine that disappears during the labeling. n = 3. (c) Testing impairment of mitochondrial 1C metabolism using serine isotope scrambing in SHMT1 KO cells. Impairments in mitochondrial 1C metabolism downstream of 13CH2-THF are reflected in increased generation of 13C1-serine (see Figure 4—figure supplement 1). n = 2. (d) Testing impairment of mitochondrial 1C metabolism by assaying formate production from serine using isolated mitochondria with acute RC inhibition. n = 3.
Figure 4—figure supplement 1. Rationale of serine isotope scrambling assay.
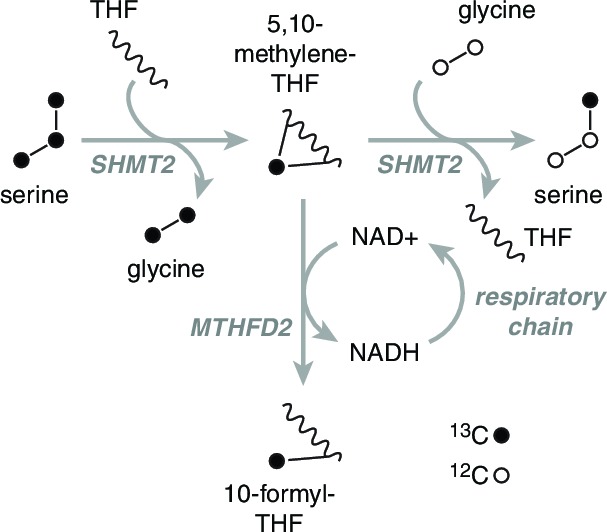
Figure 4—figure supplement 2. Alterations in cellular NAD+/NADH ratio elicited by mitochondrial manipulations.
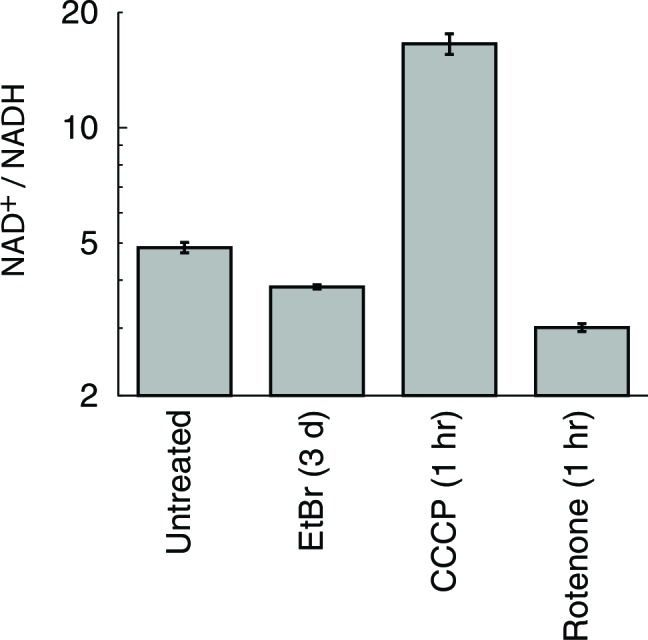
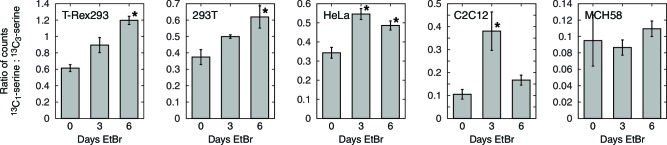
Figure 4—figure supplement 3. Serine isotope scrambling in other cell types. *, different from 0 day EtBr with p<0.05 (n = 3).
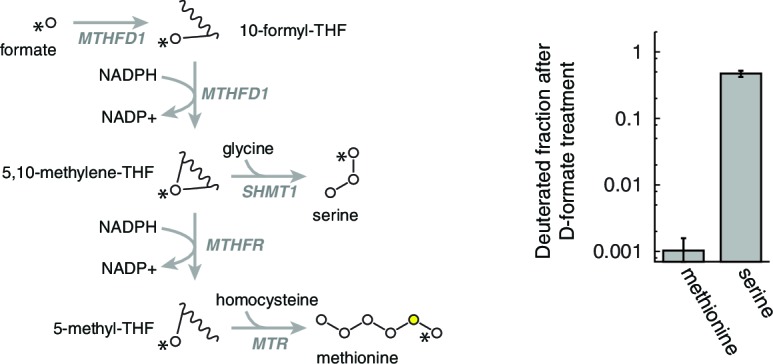
Figure 4—figure supplement 4. Determination of homocysteine remethylation.