Fig. 2.
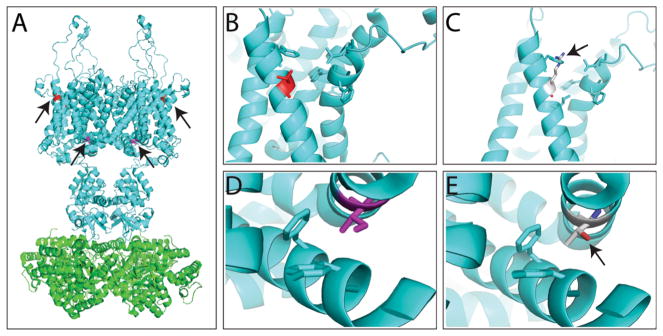
Molecular modeling of KCNA1 (KV1.1) variants. a Molecular structure of rat KCNA2 (KV1.2) (PDB code: 3LUT), a protein very similar to KCNA1, and for which crystal structure is available. The structural modeling and illustration were prepared by the software PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC). The channel forms a tetramer, with Thr226 residue in red and Ile314 colored in purple (marked by arrows). b, c WT Thr226 residue (b) and variant Thr226Arg (c) modeling. c The variant Thr226Arg conflicts with a Phe residue on the same helix (marked by arrow) and likely affects the hydrophobic packing with another helix through the interaction with the aromatic residues. d, e WT Ile314 residue (d) and variant Ile314Thr (e) modeling. e The variant Ile314Thr likely disrupts the hydrophobic packing between two transmembrane helices (marked by arrow)