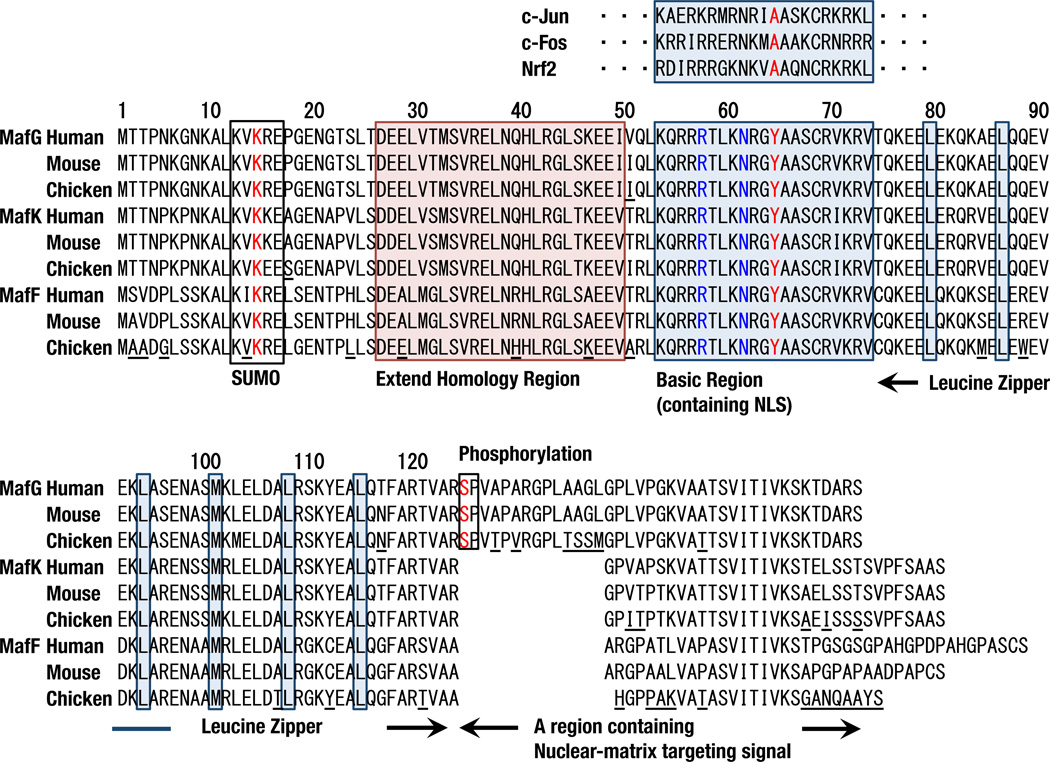
Fig. 3.
Amino acid sequence alignments of human, mouse and chicken sMafs. SUMO consensus motifs, extended homology regions, basic regions, leucine zipper leucine residues, and phosphorylation sites are indicated by boxes. Amino acid residues not conserved among individual sMafs are underlined in the chicken sequences. MafF is less conserved among species. Tyr64 in red is conserved among basic regions of the small and large Maf families but not in the other bZIP factors. Arg57 and Asn61 in blue are not unique to the Maf family but critical for GC binding.